bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4319_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
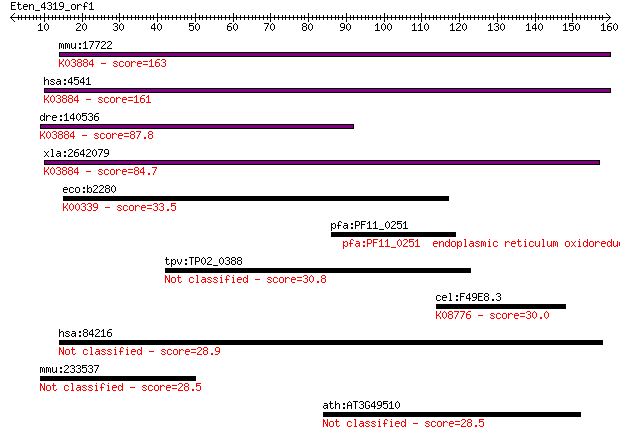
mmu:17722 ND6; NADH dehydrogenase subunit 6 (EC:1.6.5.3); K038... 163 2e-40
hsa:4541 ND6, MTND6; NADH dehydrogenase, subunit 6 (complex I)... 161 1e-39
dre:140536 ND6, mtnd6; NADH dehydrogenase subunit 6; K03884 NA... 87.8 1e-17
xla:2642079 ND6; NADH dehydrogenase subunit 6; K03884 NADH deh... 84.7 1e-16
eco:b2280 nuoJ, ECK2274, JW2275; NADH:ubiquinone oxidoreductas... 33.5 0.31
pfa:PF11_0251 endoplasmic reticulum oxidoreductin, putative; K... 31.2 1.7
tpv:TP02_0388 ABC transporter 30.8 2.2
cel:F49E8.3 pam-1; Puromycin-sensitive AMinopeptidase family m... 30.0 3.6
hsa:84216 TMEM117, DKFZp434K2435; transmembrane protein 117 28.9
mmu:233537 Gdpd4, 4921513O04; glycerophosphodiester phosphodie... 28.5 8.8
ath:AT3G49510 F-box family protein 28.5 9.8
> mmu:17722 ND6; NADH dehydrogenase subunit 6 (EC:1.6.5.3); K03884
NADH dehydrogenase I subunit 6 [EC:1.6.5.3]
Length=172
Score = 163 bits (412), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 88/146 (60%), Positives = 111/146 (76%), Gaps = 1/146 (0%)
Query 14 VFILSVIFVIGFVGFSSKPSPIYGGLGLIVSGGVGCGIVLNFGGSFLGLMVFLIYLGGMM 73
+F+LS +F++G +G + KPSPIYGGLGLIVSG VGC +VL FGGSFLGLMVFLIYLGGM+
Sbjct 5 IFVLSSLFLVGCLGLALKPSPIYGGLGLIVSGFVGCLMVLGFGGSFLGLMVFLIYLGGML 64
Query 74 VVFGYTTAMATEQYPEIWLSNKAVLGAFVTGLLMEFFMVYYVLKDKEVEVVFEFNGLGDW 133
VVFGYTTAMATE+YPE W SN +LG V G++ME F++ + EV V+ +GLGDW
Sbjct 65 VVFGYTTAMATEEYPETWGSNWLILGFLVLGVIMEVFLICVLNYYDEVGVI-NLDGLGDW 123
Query 134 VIYDTGDSGFFSEEAIGIAALYSYGT 159
++Y+ D G E IG+AA+YS T
Sbjct 124 LMYEVDDVGVMLEGGIGVAAMYSCAT 149
> hsa:4541 ND6, MTND6; NADH dehydrogenase, subunit 6 (complex
I); K03884 NADH dehydrogenase I subunit 6 [EC:1.6.5.3]
Length=174
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 92/150 (61%), Positives = 110/150 (73%), Gaps = 0/150 (0%)
Query 10 ILYIVFILSVIFVIGFVGFSSKPSPIYGGLGLIVSGGVGCGIVLNFGGSFLGLMVFLIYL 69
++Y +F+LSV V+GFVGFSSKPSPIYGGL LIVSG VGC I+LNFGG ++GLMVFLIYL
Sbjct 1 MMYALFLLSVGLVMGFVGFSSKPSPIYGGLVLIVSGVVGCVIILNFGGGYMGLMVFLIYL 60
Query 70 GGMMVVFGYTTAMATEQYPEIWLSNKAVLGAFVTGLLMEFFMVYYVLKDKEVEVVFEFNG 129
GGMMVVFGYTTAMA E+YPE W S VL + + GL ME +V +V + V VV FN
Sbjct 61 GGMMVVFGYTTAMAIEEYPEAWGSGVEVLVSVLVGLAMEVGLVLWVKEYDGVVVVVNFNS 120
Query 130 LGDWVIYDTGDSGFFSEEAIGIAALYSYGT 159
+G W+IY+ SG E+ IG ALY YG
Sbjct 121 VGSWMIYEGEGSGLIREDPIGAGALYDYGR 150
> dre:140536 ND6, mtnd6; NADH dehydrogenase subunit 6; K03884
NADH dehydrogenase I subunit 6 [EC:1.6.5.3]
Length=172
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 41/83 (49%), Positives = 61/83 (73%), Gaps = 1/83 (1%)
Query 9 MILYIVFILSVIFVIGFVGFSSKPSPIYGGLGLIVSGGVGCGIVLNFGGSFLGLMVFLIY 68
M Y+ F+++ + V G + +S P+P + GL+V GVGCGI++++GGSFL L++FLIY
Sbjct 1 MAFYLSFLMAAL-VGGMIAIASNPAPYFAAFGLVVVAGVGCGILVSYGGSFLSLILFLIY 59
Query 69 LGGMMVVFGYTTAMATEQYPEIW 91
LGGM+VVF Y+ A+A E +PE W
Sbjct 60 LGGMLVVFAYSAALAAEPFPEAW 82
> xla:2642079 ND6; NADH dehydrogenase subunit 6; K03884 NADH dehydrogenase
I subunit 6 [EC:1.6.5.3]
Length=170
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 56/147 (38%), Positives = 89/147 (60%), Gaps = 12/147 (8%)
Query 10 ILYIVFILSVIFVIGFVGFSSKPSPIYGGLGLIVSGGVGCGIVLNFGGSFLGLMVFLIYL 69
++Y+V + ++ V+G V +S PSP Y LGL+++ G GC ++++FG SFL +++FLIYL
Sbjct 1 MIYMVSVSMMVLVLGLVAVASNPSPFYAALGLVLAAGAGCLVIVSFGSSFLSIVLFLIYL 60
Query 70 GGMMVVFGYTTAMATEQYPEIWLSNKAVLGAFVTGLLMEFFMVYYVLKDKEVEVVFEFNG 129
GGM+VVF Y+ A A + YPE W S V +V L+ + Y L EV+ + + +
Sbjct 61 GGMLVVFAYSAARA-KPYPEAWGSWSVVF--YVLVYLIGVLVWYLFLGGVEVDGMNKSSE 117
Query 130 LGDWVIYDTGDSGFFSEEAIGIAALYS 156
LG +V+ GD +G+A +YS
Sbjct 118 LGSYVM--RGD-------WVGVALMYS 135
> eco:b2280 nuoJ, ECK2274, JW2275; NADH:ubiquinone oxidoreductase,
membrane subunit J; K00339 NADH dehydrogenase I subunit
J [EC:1.6.5.3]
Length=184
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 51/108 (47%), Gaps = 7/108 (6%)
Query 15 FILSVIFVIGFVGFSSKPSPIYGGLGLIVSGGVGCGIVLNFGGSFLGLMVFLIYLGGMMV 74
+I +I ++ + + +P++ L LI+S G+ + G F G + ++Y G +MV
Sbjct 6 YICGLIAILATLRVITHTNPVHALLYLIISLLAISGVFFSLGAYFAGALEIIVYAGAIMV 65
Query 75 VFGYTTAMAT------EQYPEIWLSNKAVLGAFVTGLLMEFFMVYYVL 116
+F + M EQ + WL + +G + +M +VY +L
Sbjct 66 LFVFVVMMLNLGGSEIEQERQ-WLKPQVWIGPAILSAIMLVVIVYAIL 112
> pfa:PF11_0251 endoplasmic reticulum oxidoreductin, putative;
K00540 [EC:1.-.-.-]
Length=465
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 86 QYPEIWLSNKAVLGAFVTGLLMEFFMVYYVLKD 118
QYPE +L+N+ +LG V + + + Y +LK+
Sbjct 51 QYPESYLNNEKLLGLHVRDIEKDAYRAYPILKE 83
> tpv:TP02_0388 ABC transporter
Length=1506
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 36/81 (44%), Gaps = 3/81 (3%)
Query 42 IVSGGVGCGIVLNFGGSFLGLMVFLIYLGGMMVVFGYTTAMATEQYPEIWLSNKAVLGAF 101
I+SG GCG NF S LG M + G M VV +T+ EIWL +
Sbjct 528 IISGNQGCGKS-NFIKSVLGEMTLI--EGSMAVVPLHTSMPIFYASQEIWLQQGTIRSNI 584
Query 102 VTGLLMEFFMVYYVLKDKEVE 122
+ G + + VLK E+E
Sbjct 585 IFGHKFDEVLYNTVLKAVELE 605
> cel:F49E8.3 pam-1; Puromycin-sensitive AMinopeptidase family
member (pam-1); K08776 puromycin-sensitive aminopeptidase [EC:3.4.11.-]
Length=948
Score = 30.0 bits (66), Expect = 3.6, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 114 YVLKDKEVEVVFEFNGLGDWVIYDTGDSGFFSEE 147
++LK+K+ E E G+WV ++G +GF+ E
Sbjct 583 FLLKEKQQEFTIEGVAPGEWVKLNSGTTGFYRVE 616
> hsa:84216 TMEM117, DKFZp434K2435; transmembrane protein 117
Length=514
Score = 28.9 bits (63), Expect = 8.0, Method: Composition-based stats.
Identities = 34/149 (22%), Positives = 66/149 (44%), Gaps = 19/149 (12%)
Query 14 VFILSVIFVIGFVGFSSKPSPIYGGLGLIVSGGVGCGIVLNFGG----SFLGLMVFLIYL 69
+F +++F+ F S IY + L++ G +G I+ ++ G SF+ L ++
Sbjct 111 MFFSTILFLFIF-------SHIYNTI-LLMDGNMGAYIITDYMGIRNESFMKLAAVGTWM 162
Query 70 GGMMVVFGYTTAMATEQ-YPEIWLSNKAVLGAFVTGLLMEFFMVYYVLKDKEVEVVFEFN 128
G + + T M ++ YP+ S +A + + F+ V + L V V+
Sbjct 163 GDFVTAWMVTDMMLQDKPYPDWGKSARAFWKKGNVRITL-FWTVLFTLTSVVVLVI---- 217
Query 129 GLGDWVIYDTGDSGFFSEEAIGIAALYSY 157
DW+ +D + GF + + A L S+
Sbjct 218 -TTDWISWDKLNRGFLPSDEVSRAFLASF 245
> mmu:233537 Gdpd4, 4921513O04; glycerophosphodiester phosphodiesterase
domain containing 4
Length=632
Score = 28.5 bits (62), Expect = 8.8, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 23/41 (56%), Gaps = 4/41 (9%)
Query 9 MILYIVFILSVIFVIGFVGFSSKPSPIYGGLGLIVSGGVGC 49
M L I FILSV+F+ +V S++ Y G +V G GC
Sbjct 68 MCLVIAFILSVLFLFVWVETSNE----YNGFDWVVYLGTGC 104
> ath:AT3G49510 F-box family protein
Length=662
Score = 28.5 bits (62), Expect = 9.8, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 37/88 (42%), Gaps = 20/88 (22%)
Query 84 TEQYPEIWLSNK------------AVLGAFVTGLLMEFFMVY-----YVLKDKEVEVVFE 126
T + EIW+S K V + + GL F Y ++ ++K+V V F+
Sbjct 550 TREIIEIWISTKIEPNTVSWSTFLTVDTSLINGLPDRFSTFYGPRSFFIDEEKKVAVFFD 609
Query 127 FNGLGD---WVIYDTGDSGFFSEEAIGI 151
G V Y GD+G+F IG+
Sbjct 610 NKGTETGCYQVAYIIGDNGYFKSVKIGV 637
Lambda K H
0.329 0.149 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40