bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4233_orf4
Length=233
Score E
Sequences producing significant alignments: (Bits) Value
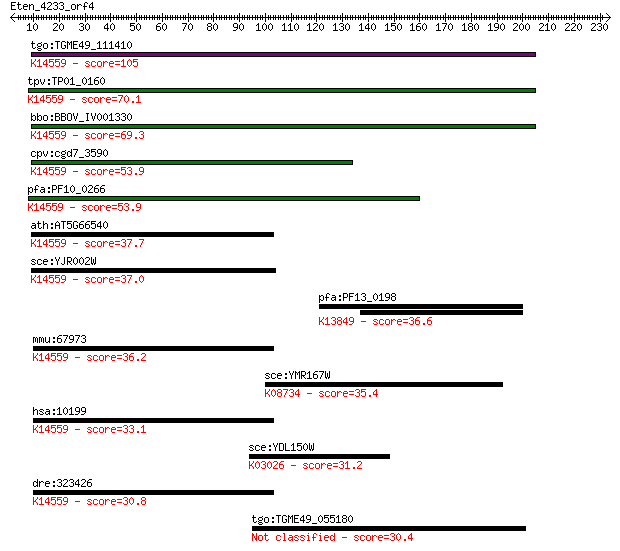
tgo:TGME49_111410 U3 small nucleolar ribonucleoprotein protein... 105 2e-22
tpv:TP01_0160 hypothetical protein; K14559 U3 small nucleolar ... 70.1 6e-12
bbo:BBOV_IV001330 21.m02752; U3 small nucleolar ribonucleoprot... 69.3 1e-11
cpv:cgd7_3590 Mpp10 family of U3 processosome protein ; K14559... 53.9 5e-07
pfa:PF10_0266 mpp10 protein, putative; K14559 U3 small nucleol... 53.9 5e-07
ath:AT5G66540 hypothetical protein; K14559 U3 small nucleolar ... 37.7 0.032
sce:YJR002W MPP10; Component of the SSU processome and 90S pre... 37.0 0.066
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 36.6 0.074
mmu:67973 Mphosph10, 2810453H10Rik, 5730405D16Rik, AI326008; M... 36.2 0.091
sce:YMR167W MLH1, PMS2; Mlh1p; K08734 DNA mismatch repair prot... 35.4 0.17
hsa:10199 MPHOSPH10, CT90, MPP10, MPP10P; M-phase phosphoprote... 33.1 0.81
sce:YDL150W RPC53, RPC4; C53; K03026 DNA-directed RNA polymera... 31.2 3.2
dre:323426 mphosph10, fb98b07, wu:fb98b07; M-phase phosphoprot... 30.8 4.2
tgo:TGME49_055180 ubiquitin carboxyl-terminal hydrolase, putat... 30.4 5.2
> tgo:TGME49_111410 U3 small nucleolar ribonucleoprotein protein
MPP10, putative ; K14559 U3 small nucleolar RNA-associated
protein MPP10
Length=866
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 79/203 (38%), Positives = 117/203 (57%), Gaps = 10/203 (4%)
Query 9 ELNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQ---LLQLFSQIMYKCDALTN 65
EL+MEK GLGD+YA +Y ++VLG + ++ E++Q LL++F +MYK D L+N
Sbjct 650 ELDMEKSKVGLGDLYAIEYEKQVLGGEAGGSGKMSAEEKQKAELLEMFGSLMYKLDCLSN 709
Query 66 YQF--VPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASSSSSSS--KAALPFTPKTAAE 121
F P D A+V+VEEA+PV + A + KA+ K E
Sbjct 710 MSFRPAPAAVAEKAKLGGGDGIAAVQVEEAVPVLLSGAMRKAPEELRKAS---KDKEKDE 766
Query 122 ATREERRAARRSKKAKRQQQLRRRIDAGELTLQQAQQRQQLLQQKNKAAKEEKKHKRLTG 181
T+ ER+A RR+KK +R++++ ++ GELT + ++R+ L KNK K K +K+ TG
Sbjct 767 MTQAERKAERRNKKERRRKRVLGQVRRGELTKEGLKERETKLAAKNKEQKANKLNKKQTG 826
Query 182 LTHAEALKELRQGQKRVKTIHLL 204
LT EAL ELR+ Q+RVK LL
Sbjct 827 LTQQEALAELRKNQRRVKVQELL 849
> tpv:TP01_0160 hypothetical protein; K14559 U3 small nucleolar
RNA-associated protein MPP10
Length=576
Score = 70.1 bits (170), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 54/197 (27%), Positives = 102/197 (51%), Gaps = 25/197 (12%)
Query 8 EELNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNYQ 67
EEL+ K GLG+IY +Y++ S +LD +KQ L Q+FS++MYK D+L+N
Sbjct 399 EELDFSKDKRGLGEIYEDKYKQLF----SSNNSKLDAQKQLLTQMFSKLMYKLDSLSNCS 454
Query 68 FVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASSSSSSSKAALPFTPKTAAEATREER 127
FVP+ +++ VE P+ V + ++++++K ++
Sbjct 455 FVPKIPKITTNNV-----STISVE--TPINVIISENNTNTNK---------KVDSNLISV 498
Query 128 RAARRSKKAKRQQQLRRRIDAGELTLQQAQQRQQLLQQKNKAAKEEKKHKRLTGLTHAEA 187
R KK K + +++R + +G+++ Q+ + + +++ NK +EE K+K+LTGL +
Sbjct 499 STNRNRKKRKFKSKMKRLVKSGKMSAQEVEMLNKKMRETNKKRREELKNKKLTGLPGKDI 558
Query 188 LKELRQGQKRVKTIHLL 204
+ + R+ T LL
Sbjct 559 -----KSRTRINTTQLL 570
> bbo:BBOV_IV001330 21.m02752; U3 small nucleolar ribonucleoprotein
protein; K14559 U3 small nucleolar RNA-associated protein
MPP10
Length=653
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 53/196 (27%), Positives = 95/196 (48%), Gaps = 24/196 (12%)
Query 9 ELNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNYQF 68
E++ K GLGD+YAQ+Y+E + +LD KQ+L + F +IM+K D+L NY
Sbjct 478 EIDFNKSKMGLGDVYAQRYQEMFMAT-----DKLDAHKQKLAEDFGKIMFKLDSLCNYHI 532
Query 69 VPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASSSSSSSKAALPFTPKTAAEATREERR 128
VP+ AS+ VE I V VPA+ + + E +
Sbjct 533 VPKRVSEAEKNKNV---ASITVEAPINV-VPASRHNDVTD--------------ISEGNK 574
Query 129 AARRSKKAKRQQQLRRRIDAGELTLQQAQQRQQLLQQKNKAAKEEKKHKRLTGLTHAEAL 188
+R +KK K +++ + +G+ T++ ++ +Q + ++N+ E+ + + TG T
Sbjct 575 LSRTAKKRKFTNKIKALLKSGKATVEDIKRMKQKMVERNRQKIEDMRSVKATGQTKERGA 634
Query 189 KELRQGQKRVKTIHLL 204
+E ++ RVK L+
Sbjct 635 RE-QKPSGRVKIAELM 649
> cpv:cgd7_3590 Mpp10 family of U3 processosome protein ; K14559
U3 small nucleolar RNA-associated protein MPP10
Length=551
Score = 53.9 bits (128), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 38/129 (29%), Positives = 66/129 (51%), Gaps = 13/129 (10%)
Query 9 ELNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNYQF 68
E+ EK L +IY+++Y E+++G + + EK+QL +LF++IM K D L+N +
Sbjct 375 EIQTEKSKLSLAEIYSKKYEEQIIGNINDEHSD---EKKQLSELFTEIMRKLDNLSNQYY 431
Query 69 VPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASSSSSSSKAALPFTPK----TAAEATR 124
+ PA ++ E++IPV + S+S+ A P+ +E TR
Sbjct 432 ATNLPILRDSILNENTPA-LKTEDSIPVII-----SNSTRLAPQEIKPQGRLLKRSEMTR 485
Query 125 EERRAARRS 133
E+R+ R S
Sbjct 486 LEKRSDRNS 494
> pfa:PF10_0266 mpp10 protein, putative; K14559 U3 small nucleolar
RNA-associated protein MPP10
Length=772
Score = 53.9 bits (128), Expect = 5e-07, Method: Composition-based stats.
Identities = 39/155 (25%), Positives = 84/155 (54%), Gaps = 11/155 (7%)
Query 8 EELNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNYQ 67
+ LN K L D Y Q+Y+++++ + S+ ++ + +K +++ LF +IM+ CD+L+N
Sbjct 569 DNLNFSKSKISLVDEYTQKYQDEIMNSQSKNKEN-NLQKMEIMNLFKKIMHACDSLSNDY 627
Query 68 FVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASSSSSSSKAALPFTP---KTAAEATR 124
F+P+ A++ +E+++P+ + S + A F P K E T+
Sbjct 628 FIPKPILLNNKNNKM---ATLNIEDSVPMIL----SDKNKKTAEEIFNPQHMKQQNELTK 680
Query 125 EERRAARRSKKAKRQQQLRRRIDAGELTLQQAQQR 159
+E++A R+SKK K+++++ + ++ Q +R
Sbjct 681 DEKKAIRKSKKMKKKKKILNQFKQTGSSINQLVKR 715
> ath:AT5G66540 hypothetical protein; K14559 U3 small nucleolar
RNA-associated protein MPP10
Length=524
Score = 37.7 bits (86), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 46/96 (47%), Gaps = 8/96 (8%)
Query 9 ELNMEKPAEGLGDIYAQQYREKV--LGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNY 66
EL+ K +GL ++Y +Y +K AP+ +L K++ LF ++ K DAL+++
Sbjct 379 ELDESKSKKGLAEVYEAEYFQKANPAFAPTTHSDEL---KKEASMLFKKLCLKLDALSHF 435
Query 67 QFVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAAS 102
F P+ A + +EE PV V A+
Sbjct 436 HFTPKPVIEEMSIPNVSA---IAMEEVAPVAVSDAA 468
> sce:YJR002W MPP10; Component of the SSU processome and 90S preribosome,
required for pre-18S rRNA processing, interacts
with and controls the stability of Imp3p and Imp4p, essential
for viability; similar to human Mpp10p; K14559 U3 small nucleolar
RNA-associated protein MPP10
Length=593
Score = 37.0 bits (84), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 52/96 (54%), Gaps = 6/96 (6%)
Query 9 ELNMEKPAEGLGDIYAQQY-REKVLGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNYQ 67
EL+ K ++ L +IY Y R + A S++ Q+ E + +L++ ++YK D L++
Sbjct 399 ELSDVKSSKSLAEIYEDDYTRAEDESALSEELQKAHSE---ISELYANLVYKLDVLSSVH 455
Query 68 FVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAASS 103
FVP+ + P ++ +E+A P+Y+ ASS
Sbjct 456 FVPK-PASTSLEIRVETP-TISMEDAQPLYMSNASS 489
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 36.6 bits (83), Expect = 0.074, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 42/79 (53%), Gaps = 4/79 (5%)
Query 121 EATREERRAARRSKKAKRQQQLRRRIDAGELTLQQAQQRQQLLQQKNKAAKEEKKHKRLT 180
E R+E+ ++ + KRQ+Q R+ E +Q Q+R + +Q+ +EE K +
Sbjct 2774 ELKRQEQERLQKEEALKRQEQ--ERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQE 2831
Query 181 GLTHAEALKELRQGQKRVK 199
L EALK RQ Q+R++
Sbjct 2832 RLQKEEALK--RQEQERLQ 2848
Score = 30.8 bits (68), Expect = 4.2, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Query 137 KRQQQLRRRIDAGELTLQQAQQRQQLLQQKNKAAKEEKKHKRLTGLTHAEALKELRQGQK 196
KRQ+Q R+ E +Q Q+R + +Q+ +EE K + L EALK RQ Q+
Sbjct 2740 KRQEQ--ERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALK--RQEQE 2795
Query 197 RVK 199
R++
Sbjct 2796 RLQ 2798
> mmu:67973 Mphosph10, 2810453H10Rik, 5730405D16Rik, AI326008;
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein);
K14559 U3 small nucleolar RNA-associated protein MPP10
Length=681
Score = 36.2 bits (82), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 46/94 (48%), Gaps = 7/94 (7%)
Query 10 LNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQ-EKQQLLQLFSQIMYKCDALTNYQF 68
L+ EK L +IY Q+Y L Q+ ++ D E ++ ++ + K DAL+N+ F
Sbjct 461 LDHEKSKLSLAEIYEQEY----LKLNQQKTEEEDNPEHVEIQKMMDSLFLKLDALSNFHF 516
Query 69 VPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAAS 102
+P+ + PA + +EE PV V A+
Sbjct 517 IPK-PPVPEIKVVSNLPA-ITMEEVAPVSVSDAA 548
> sce:YMR167W MLH1, PMS2; Mlh1p; K08734 DNA mismatch repair protein
MLH1
Length=769
Score = 35.4 bits (80), Expect = 0.17, Method: Composition-based stats.
Identities = 22/103 (21%), Positives = 48/103 (46%), Gaps = 11/103 (10%)
Query 100 AASSSSSSSKAALPFTPKTAAEATREERRAAR-----------RSKKAKRQQQLRRRIDA 148
A+S S++ ++ +PF ++ R+ R A+ + +KAKRQ+ RIDA
Sbjct 345 ASSISTNKPESLIPFNDTIESDRNRKSLRQAQVVENSYTTANSQLRKAKRQENKLVRIDA 404
Query 149 GELTLQQAQQRQQLLQQKNKAAKEEKKHKRLTGLTHAEALKEL 191
+ + Q + + K + ++T ++H++ ++L
Sbjct 405 SQAKITSFLSSSQQFNFEGSSTKRQLSEPKVTNVSHSQEAEKL 447
> hsa:10199 MPHOSPH10, CT90, MPP10, MPP10P; M-phase phosphoprotein
10 (U3 small nucleolar ribonucleoprotein); K14559 U3 small
nucleolar RNA-associated protein MPP10
Length=681
Score = 33.1 bits (74), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 11/96 (11%)
Query 10 LNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQLLQ---LFSQIMYKCDALTNY 66
L+ EK L +IY Q+Y QQ+ ++E + ++ + + K DAL+N+
Sbjct 460 LDHEKSKLSLAEIYEQEY------IKLNQQKTAEEENPEHVEIQKMMDSLFLKLDALSNF 513
Query 67 QFVPRXXXXXXXXXXXDAPASVRVEEAIPVYVPAAS 102
F+P+ + PA + +EE PV V A+
Sbjct 514 HFIPK-PPVPEIKVVSNLPA-ITMEEVAPVSVSDAA 547
> sce:YDL150W RPC53, RPC4; C53; K03026 DNA-directed RNA polymerase
III subunit RPC4
Length=422
Score = 31.2 bits (69), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 31/54 (57%), Gaps = 2/54 (3%)
Query 94 IPVYVPAASSSSSSSKAALPFTPKTAAEATREERRAARRSKKAKRQQQLRRRID 147
+P ++S+ S+K +L F PK A ++EER AA + K K +++ +R D
Sbjct 10 LPSLKDSSSNGGGSAKPSLKFKPKAVARKSKEEREAA--ASKVKLEEESKRGND 61
> dre:323426 mphosph10, fb98b07, wu:fb98b07; M-phase phosphoprotein
10 (U3 small nucleolar ribonucleoprotein); K14559 U3 small
nucleolar RNA-associated protein MPP10
Length=695
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 24/93 (25%), Positives = 44/93 (47%), Gaps = 5/93 (5%)
Query 10 LNMEKPAEGLGDIYAQQYREKVLGAPSQQQQQLDQEKQQLLQLFSQIMYKCDALTNYQFV 69
L+ EK L ++Y Q+Y ++ +++ E Q+L+ + K DAL+N+ F
Sbjct 470 LDHEKSKLSLAEVYEQEYIKQTQEKKEEEENPAHVEIQKLM---DSLFLKLDALSNFHFT 526
Query 70 PRXXXXXXXXXXXDAPASVRVEEAIPVYVPAAS 102
P+ + P ++ +EE PV V A+
Sbjct 527 PK-PHVPEVKVVSNLP-TITMEEVAPVNVSDAT 557
> tgo:TGME49_055180 ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15)
Length=4187
Score = 30.4 bits (67), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 30/117 (25%), Positives = 53/117 (45%), Gaps = 11/117 (9%)
Query 95 PVYVPAASSSSSSSKAALPFTPKTAAEATRE---------ERRAARRSKKAKRQQQLRRR 145
P +P S++ + +P T + R E+R A S + K + +LR R
Sbjct 2424 PSLLPRRDSNNRNGICMVPLTKAENGDGNRPARKRKKNNAEQRGAAGSSREKSEDELRER 2483
Query 146 IDAGELTLQQAQQRQQLLQQK-NKAAKEEKKHKRLTGLTHAEALK-ELRQGQKRVKT 200
G + A + L+ N AK+E K + L GL+ E L+ E R+G+++ ++
Sbjct 2484 GRLGAPGEEHATAGESELEGSANAGAKKEAKEEDLRGLSICEVLEGETREGRQQSRS 2540
Lambda K H
0.314 0.125 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7953524176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40