bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4181_orf2
Length=212
Score E
Sequences producing significant alignments: (Bits) Value
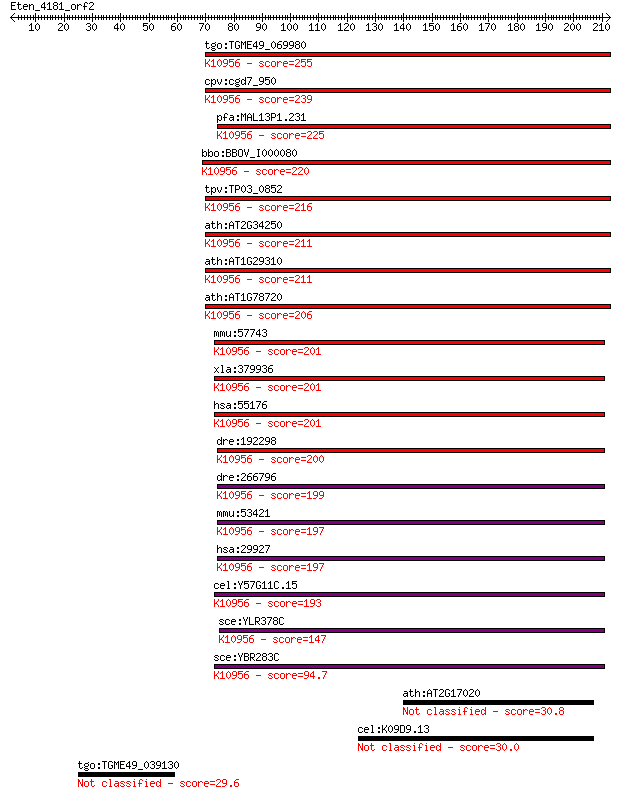
tgo:TGME49_069980 protein transport protein Sec61 alpha subuni... 255 9e-68
cpv:cgd7_950 Sec61; signal peptide plus 9 transmembrane domain... 239 5e-63
pfa:MAL13P1.231 sec61, PF13_0244; Sec61 alpha subunit, PfSec61... 225 1e-58
bbo:BBOV_I000080 16.m00785; protein transport protein sec61; K... 220 2e-57
tpv:TP03_0852 protein transport protein Sec61; K10956 protein ... 216 4e-56
ath:AT2G34250 protein transport protein sec61, putative; K1095... 211 1e-54
ath:AT1G29310 P-P-bond-hydrolysis-driven protein transmembrane... 211 2e-54
ath:AT1G78720 protein transport protein sec61, putative; K1095... 206 4e-53
mmu:57743 Sec61a2; Sec61, alpha subunit 2 (S. cerevisiae); K10... 201 1e-51
xla:379936 sec61a1, MGC53497, sec61, sec61a, sec61alpha; Sec61... 201 1e-51
hsa:55176 SEC61A2, FLJ10578; Sec61 alpha 2 subunit (S. cerevis... 201 1e-51
dre:192298 sec61al1, chunp6898, fb62c11, sec61a, wu:fb62c11; S... 200 3e-51
dre:266796 sec61al2, cb413, sec61b, wu:fb36f05, wu:fb36f12, wu... 199 7e-51
mmu:53421 Sec61a1, AA408394, AA410007, Sec61a; Sec61 alpha 1 s... 197 2e-50
hsa:29927 SEC61A1, HSEC61, SEC61, SEC61A; Sec61 alpha 1 subuni... 197 2e-50
cel:Y57G11C.15 hypothetical protein; K10956 protein transport ... 193 3e-49
sce:YLR378C SEC61; Essential subunit of Sec61 complex (Sec61p,... 147 4e-35
sce:YBR283C SSH1; Ssh1p; K10956 protein transport protein SEC6... 94.7 2e-19
ath:AT2G17020 F-box family protein (FBL10) 30.8 3.3
cel:K09D9.13 srw-89; Serpentine Receptor, class W family membe... 30.0 6.0
tgo:TGME49_039130 protein kinase, putative (EC:2.7.10.2) 29.6 8.9
> tgo:TGME49_069980 protein transport protein Sec61 alpha subunit
isoform 1, putative ; K10956 protein transport protein SEC61
subunit alpha
Length=473
Score = 255 bits (651), Expect = 9e-68, Method: Compositional matrix adjust.
Identities = 117/143 (81%), Positives = 133/143 (93%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
MAKG RFL+L+KP MC+LPEV AP+R++ F+EKVLWTL++LAVFL+CCQIPL+GIR +
Sbjct 1 MAKGFRFLNLIKPVMCILPEVQAPDRKIPFKEKVLWTLVSLAVFLICCQIPLYGIRTNKS 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSL+EDRALFQGAQK
Sbjct 61 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLKEDRALFQGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL L+IT+ EAVAYVISGMYG +
Sbjct 121 LLGLIITVGEAVAYVISGMYGDI 143
> cpv:cgd7_950 Sec61; signal peptide plus 9 transmembrane domain-containing
protein ; K10956 protein transport protein SEC61
subunit alpha
Length=473
Score = 239 bits (610), Expect = 5e-63, Method: Compositional matrix adjust.
Identities = 110/143 (76%), Positives = 130/143 (90%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
MAKG+RFL+L+KPA+CV+PEV +P+RRV F+E+VLWTL++L VFLVCCQIPL+G+ +
Sbjct 1 MAKGLRFLNLIKPALCVIPEVSSPDRRVPFKERVLWTLISLFVFLVCCQIPLYGVLSSKS 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
+DPFYW+RVILASNRGTLMELGISPIVTS MVMQLLAGS+II VDQSL+EDRALFQGAQK
Sbjct 61 SDPFYWVRVILASNRGTLMELGISPIVTSSMVMQLLAGSKIIDVDQSLKEDRALFQGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
L LLIT+ EAVAYVISGMYG +
Sbjct 121 LFGLLITLGEAVAYVISGMYGDI 143
> pfa:MAL13P1.231 sec61, PF13_0244; Sec61 alpha subunit, PfSec61;
K10956 protein transport protein SEC61 subunit alpha
Length=472
Score = 225 bits (573), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 102/139 (73%), Positives = 124/139 (89%), Gaps = 0/139 (0%)
Query 74 IRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPF 133
+RFL+L+KP M +LPEV +P+R++ F+EK+LWT ++L VFL+CCQIPL+GI + +DPF
Sbjct 2 VRFLNLIKPVMFLLPEVQSPDRKLPFKEKLLWTAVSLFVFLICCQIPLYGIVTSKSSDPF 61
Query 134 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLAL 193
YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGS+II VDQSL+EDR LFQGAQKLL L
Sbjct 62 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSKIIDVDQSLKEDRTLFQGAQKLLGL 121
Query 194 LITIXEAVAYVISGMYGPV 212
LIT+ EA+AYVISG+YG +
Sbjct 122 LITLGEAIAYVISGIYGNI 140
> bbo:BBOV_I000080 16.m00785; protein transport protein sec61;
K10956 protein transport protein SEC61 subunit alpha
Length=480
Score = 220 bits (561), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 101/144 (70%), Positives = 122/144 (84%), Gaps = 0/144 (0%)
Query 69 KMAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGR 128
KMAKG RFL+L+KP M +LPE+ P R+V F+E ++WT ++L VFLVCCQIP++G +
Sbjct 4 KMAKGFRFLNLIKPIMPILPEIRTPTRKVPFKEMLMWTGVSLFVFLVCCQIPIYGAVTNK 63
Query 129 GADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQ 188
+DPFYWMRVILASNRGTLMELGISPIVTS MVMQLLAGS+II VDQSL+EDR L+Q A+
Sbjct 64 SSDPFYWMRVILASNRGTLMELGISPIVTSSMVMQLLAGSKIIDVDQSLKEDRDLYQAAE 123
Query 189 KLLALLITIXEAVAYVISGMYGPV 212
KL LL+T+ EAVAYV+SGMYGPV
Sbjct 124 KLFGLLVTLGEAVAYVVSGMYGPV 147
> tpv:TP03_0852 protein transport protein Sec61; K10956 protein
transport protein SEC61 subunit alpha
Length=476
Score = 216 bits (551), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 99/143 (69%), Positives = 120/143 (83%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
M KG R L+L+KP M +LPEV P R+VLF+E ++WT ++L +FLVCCQIP++G +
Sbjct 1 MGKGFRVLNLIKPIMPILPEVKTPTRKVLFKEMLMWTGMSLFIFLVCCQIPIYGAITNKS 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
+DPFYWMRVILASNRGTLMELGISPIVTS MVMQLLAGS+II VDQSL+EDR L+Q A+K
Sbjct 61 SDPFYWMRVILASNRGTLMELGISPIVTSSMVMQLLAGSKIIDVDQSLKEDRDLYQAAEK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL LL+T+ EAVAYV+SGMYG V
Sbjct 121 LLGLLVTLGEAVAYVVSGMYGDV 143
> ath:AT2G34250 protein transport protein sec61, putative; K10956
protein transport protein SEC61 subunit alpha
Length=475
Score = 211 bits (538), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 98/143 (68%), Positives = 120/143 (83%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
M G R L LV+P + LPEV + +R+V FREKV++T+++L +FLVC Q+PL+GI G
Sbjct 1 MGGGFRVLHLVRPFLAFLPEVQSADRKVPFREKVIYTVISLFIFLVCSQLPLYGIHSTTG 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYWMRVILASNRGT+MELGI+PIVTSG+VMQLLAGS+II+VD ++REDRAL GAQK
Sbjct 61 ADPFYWMRVILASNRGTVMELGITPIVTSGLVMQLLAGSKIIEVDNNVREDRALLNGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL +LI I EAVAYV+SGMYGPV
Sbjct 121 LLGILIAIGEAVAYVLSGMYGPV 143
> ath:AT1G29310 P-P-bond-hydrolysis-driven protein transmembrane
transporter; K10956 protein transport protein SEC61 subunit
alpha
Length=475
Score = 211 bits (537), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 97/143 (67%), Positives = 120/143 (83%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
M G R L LV+P + LPEV + +R++ FREKV++T+++L +FLVC Q+PL+GI G
Sbjct 1 MGGGFRVLHLVRPFLAFLPEVQSADRKIPFREKVIYTVISLFIFLVCSQLPLYGIHSTTG 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYWMRVILASNRGT+MELGI+PIVTSG+VMQLLAGS+II+VD ++REDRAL GAQK
Sbjct 61 ADPFYWMRVILASNRGTVMELGITPIVTSGLVMQLLAGSKIIEVDNNVREDRALLNGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL +LI I EAVAYV+SGMYGPV
Sbjct 121 LLGILIAIGEAVAYVLSGMYGPV 143
> ath:AT1G78720 protein transport protein sec61, putative; K10956
protein transport protein SEC61 subunit alpha
Length=475
Score = 206 bits (525), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 94/143 (65%), Positives = 120/143 (83%), Gaps = 0/143 (0%)
Query 70 MAKGIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG 129
M G R + LV+P + LPEV +PER++ FREKV++T+++L +FLVC Q+PL+GI G
Sbjct 1 MVGGFRVIHLVRPFLAFLPEVQSPERKIPFREKVIYTVISLFIFLVCSQLPLYGIHSTTG 60
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
ADPFYWMRVILAS+RGT+MELGI+PIVTSGMVMQLLAGS+II++D ++REDRAL GAQK
Sbjct 61 ADPFYWMRVILASSRGTVMELGITPIVTSGMVMQLLAGSKIIEIDNNVREDRALLNGAQK 120
Query 190 LLALLITIXEAVAYVISGMYGPV 212
LL +LI + +AVAYV+SGMYG V
Sbjct 121 LLGILIAVGQAVAYVLSGMYGSV 143
> mmu:57743 Sec61a2; Sec61, alpha subunit 2 (S. cerevisiae); K10956
protein transport protein SEC61 subunit alpha
Length=476
Score = 201 bits (512), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 93/138 (67%), Positives = 114/138 (82%), Gaps = 1/138 (0%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADP 132
GI+FL ++KP VLPE+ PER++ FREKVLWT + L +FLVCCQIPLFGI ADP
Sbjct 2 GIKFLEVIKPFCAVLPEIQKPERKIQFREKVLWTAITLFIFLVCCQIPLFGIMSSDSADP 61
Query 133 FYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLA 192
FYWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + +DRALF GAQKL
Sbjct 62 FYWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDT-PKDRALFNGAQKLFG 120
Query 193 LLITIXEAVAYVISGMYG 210
++ITI +A+ YV++GMYG
Sbjct 121 MIITIGQAIVYVMTGMYG 138
> xla:379936 sec61a1, MGC53497, sec61, sec61a, sec61alpha; Sec61
alpha 1 subunit; K10956 protein transport protein SEC61 subunit
alpha
Length=476
Score = 201 bits (511), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 92/138 (66%), Positives = 114/138 (82%), Gaps = 1/138 (0%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADP 132
GI+FL ++KP VLPE+ PER++ F+EKVLWT + L +FLVCCQIPLFGI ADP
Sbjct 2 GIKFLEVIKPFCAVLPEIQKPERKIQFKEKVLWTAITLFIFLVCCQIPLFGIMSSDSADP 61
Query 133 FYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLA 192
FYWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL
Sbjct 62 FYWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFG 120
Query 193 LLITIXEAVAYVISGMYG 210
++ITI +A+ YV++GMYG
Sbjct 121 MIITIGQAIVYVMTGMYG 138
> hsa:55176 SEC61A2, FLJ10578; Sec61 alpha 2 subunit (S. cerevisiae);
K10956 protein transport protein SEC61 subunit alpha
Length=437
Score = 201 bits (511), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 93/138 (67%), Positives = 114/138 (82%), Gaps = 1/138 (0%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADP 132
GI+FL ++KP VLPE+ PER++ FREKVLWT + L +FLVCCQIPLFGI ADP
Sbjct 2 GIKFLEVIKPFCAVLPEIQKPERKIQFREKVLWTAITLFIFLVCCQIPLFGIMSSDSADP 61
Query 133 FYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLA 192
FYWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL
Sbjct 62 FYWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFG 120
Query 193 LLITIXEAVAYVISGMYG 210
++ITI +A+ YV++GMYG
Sbjct 121 MIITIGQAIVYVMTGMYG 138
> dre:192298 sec61al1, chunp6898, fb62c11, sec61a, wu:fb62c11;
Sec61 alpha like 1; K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 200 bits (508), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 92/137 (67%), Positives = 113/137 (82%), Gaps = 1/137 (0%)
Query 74 IRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPF 133
I+FL ++KP VLPE+ PER++ FREKVLWT + L +FLVCCQIPLFGI ADPF
Sbjct 3 IKFLEVIKPFCAVLPEIQKPERKIQFREKVLWTAITLFIFLVCCQIPLFGIMSSDSADPF 62
Query 134 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLAL 193
YWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL +
Sbjct 63 YWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFGM 121
Query 194 LITIXEAVAYVISGMYG 210
+ITI +A+ YV++GMYG
Sbjct 122 IITIGQAIVYVMTGMYG 138
> dre:266796 sec61al2, cb413, sec61b, wu:fb36f05, wu:fb36f12,
wu:fb37d04, wu:fi26f11; Sec61 alpha like 2; K10956 protein transport
protein SEC61 subunit alpha
Length=476
Score = 199 bits (505), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 91/137 (66%), Positives = 113/137 (82%), Gaps = 1/137 (0%)
Query 74 IRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPF 133
I+FL ++KP VLPE+ PERR+ F+EKVLWT + L +FLVCCQIPLFGI ADPF
Sbjct 3 IKFLEVIKPFCAVLPEIQKPERRIQFKEKVLWTAITLFIFLVCCQIPLFGIMSSDSADPF 62
Query 134 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLAL 193
YWMRVI+ASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL +
Sbjct 63 YWMRVIMASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFGM 121
Query 194 LITIXEAVAYVISGMYG 210
+ITI +A+ YV++GMYG
Sbjct 122 IITIGQAIVYVMTGMYG 138
> mmu:53421 Sec61a1, AA408394, AA410007, Sec61a; Sec61 alpha 1
subunit (S. cerevisiae); K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 197 bits (502), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 113/137 (82%), Gaps = 1/137 (0%)
Query 74 IRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPF 133
I+FL ++KP +LPE+ PER++ F+EKVLWT + L +FLVCCQIPLFGI ADPF
Sbjct 3 IKFLEVIKPFCVILPEIQKPERKIQFKEKVLWTAITLFIFLVCCQIPLFGIMSSDSADPF 62
Query 134 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLAL 193
YWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL +
Sbjct 63 YWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFGM 121
Query 194 LITIXEAVAYVISGMYG 210
+ITI +++ YV++GMYG
Sbjct 122 IITIGQSIVYVMTGMYG 138
> hsa:29927 SEC61A1, HSEC61, SEC61, SEC61A; Sec61 alpha 1 subunit
(S. cerevisiae); K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 197 bits (502), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 113/137 (82%), Gaps = 1/137 (0%)
Query 74 IRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPF 133
I+FL ++KP +LPE+ PER++ F+EKVLWT + L +FLVCCQIPLFGI ADPF
Sbjct 3 IKFLEVIKPFCVILPEIQKPERKIQFKEKVLWTAITLFIFLVCCQIPLFGIMSSDSADPF 62
Query 134 YWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLAL 193
YWMRVILASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL +
Sbjct 63 YWMRVILASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFGM 121
Query 194 LITIXEAVAYVISGMYG 210
+ITI +++ YV++GMYG
Sbjct 122 IITIGQSIVYVMTGMYG 138
> cel:Y57G11C.15 hypothetical protein; K10956 protein transport
protein SEC61 subunit alpha
Length=473
Score = 193 bits (491), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 90/138 (65%), Positives = 112/138 (81%), Gaps = 1/138 (0%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADP 132
GI+FL VKP +PEV PER++ FREK+LWT + L VFLVCCQIPLFGI ADP
Sbjct 2 GIKFLEFVKPFCGFVPEVSKPERKIQFREKMLWTAITLFVFLVCCQIPLFGIMSTDSADP 61
Query 133 FYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLA 192
FYW+RVI+ASNRGTLMELGISPIVTSG++MQLLAG++II+V + + DRALF GAQKL
Sbjct 62 FYWLRVIMASNRGTLMELGISPIVTSGLIMQLLAGAKIIEVGDTPK-DRALFNGAQKLFG 120
Query 193 LLITIXEAVAYVISGMYG 210
++IT+ +A+ YV+SG+YG
Sbjct 121 MVITVGQAIVYVMSGLYG 138
> sce:YLR378C SEC61; Essential subunit of Sec61 complex (Sec61p,
Sbh1p, and Sss1p); forms a channel for SRP-dependent protein
import and retrograde transport of misfolded proteins out
of the ER; with Sec63 complex allows SRP-independent protein
import into ER; K10956 protein transport protein SEC61 subunit
alpha
Length=480
Score = 147 bits (370), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 66/136 (48%), Positives = 99/136 (72%), Gaps = 0/136 (0%)
Query 75 RFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRGADPFY 134
R L L KP LPEV APER+V + +K++WT ++L +FL+ QIPL+GI +DP Y
Sbjct 5 RVLDLFKPFESFLPEVIAPERKVPYNQKLIWTGVSLLIFLILGQIPLYGIVSSETSDPLY 64
Query 135 WMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLALL 194
W+R +LASNRGTL+ELG+SPI+TS M+ Q L G++++Q+ ++DR LFQ AQK+ A++
Sbjct 65 WLRAMLASNRGTLLELGVSPIITSSMIFQFLQGTQLLQIRPESKQDRELFQIAQKVCAII 124
Query 195 ITIXEAVAYVISGMYG 210
+ + +A+ V++G YG
Sbjct 125 LILGQALVVVMTGNYG 140
> sce:YBR283C SSH1; Ssh1p; K10956 protein transport protein SEC61
subunit alpha
Length=490
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 46/141 (32%), Positives = 84/141 (59%), Gaps = 4/141 (2%)
Query 73 GIRFLSLVKPAMCVLPEVHAPERRVLFREKVLWTLLALAVFLVCCQIPLFGIRGGRG--- 129
G R + +VKP + +LPEV P ++ F +K+++T+ A ++L Q PL G+
Sbjct 3 GFRLIDIVKPILPILPEVELPFEKLPFDDKIVYTIFAGLIYLFA-QFPLVGLPKATTPNV 61
Query 130 ADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQK 189
DP Y++R + TL+E G+ P ++SG+++QLLAG ++I+V+ ++ DR LFQ K
Sbjct 62 NDPIYFLRGVFGCEPRTLLEFGLFPNISSGLILQLLAGLKVIKVNFKIQSDRELFQSLTK 121
Query 190 LLALLITIXEAVAYVISGMYG 210
+ A++ + ++ +G +G
Sbjct 122 VFAIVQYVILTNIFIFAGYFG 142
> ath:AT2G17020 F-box family protein (FBL10)
Length=656
Score = 30.8 bits (68), Expect = 3.3, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 33/67 (49%), Gaps = 3/67 (4%)
Query 140 LASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRALFQGAQKLLALLITIXE 199
L S+R T +ELG +TS M+ QLL + I D + + Q Q+L + I +
Sbjct 192 LVSDRLTHLELGH---ITSRMMTQLLTSTEISGQDSNRVTTSTVLQNVQRLRLSVDCITD 248
Query 200 AVAYVIS 206
AV IS
Sbjct 249 AVVKAIS 255
> cel:K09D9.13 srw-89; Serpentine Receptor, class W family member
(srw-89)
Length=361
Score = 30.0 bits (66), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 40/83 (48%), Gaps = 2/83 (2%)
Query 124 IRGGRGADPFYWMRVILASNRGTLMELGISPIVTSGMVMQLLAGSRIIQVDQSLREDRAL 183
+ G + F W + A R ME S ++T + +L+G I ++ +S
Sbjct 197 VHGQVSNEIFEWNNALFAKLR-LYMECVFSKLITC-FALPILSGLLITEMKKSAEVSIKS 254
Query 184 FQGAQKLLALLITIXEAVAYVIS 206
F+ ++K L +ITI A++Y +S
Sbjct 255 FKKSRKDLTTVITICIAISYFVS 277
> tgo:TGME49_039130 protein kinase, putative (EC:2.7.10.2)
Length=1658
Score = 29.6 bits (65), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 25 WRRAAAQRSSSSSSSGAAQQQLAGRSACAPPSSL 58
W R AQ S+SS SG AQ+ AGRS AP L
Sbjct 1216 WVRQHAQNSTSSRFSGEAQKPKAGRSFAAPGCEL 1249
Lambda K H
0.325 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6687805280
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40