bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4098_orf1
Length=62
Score E
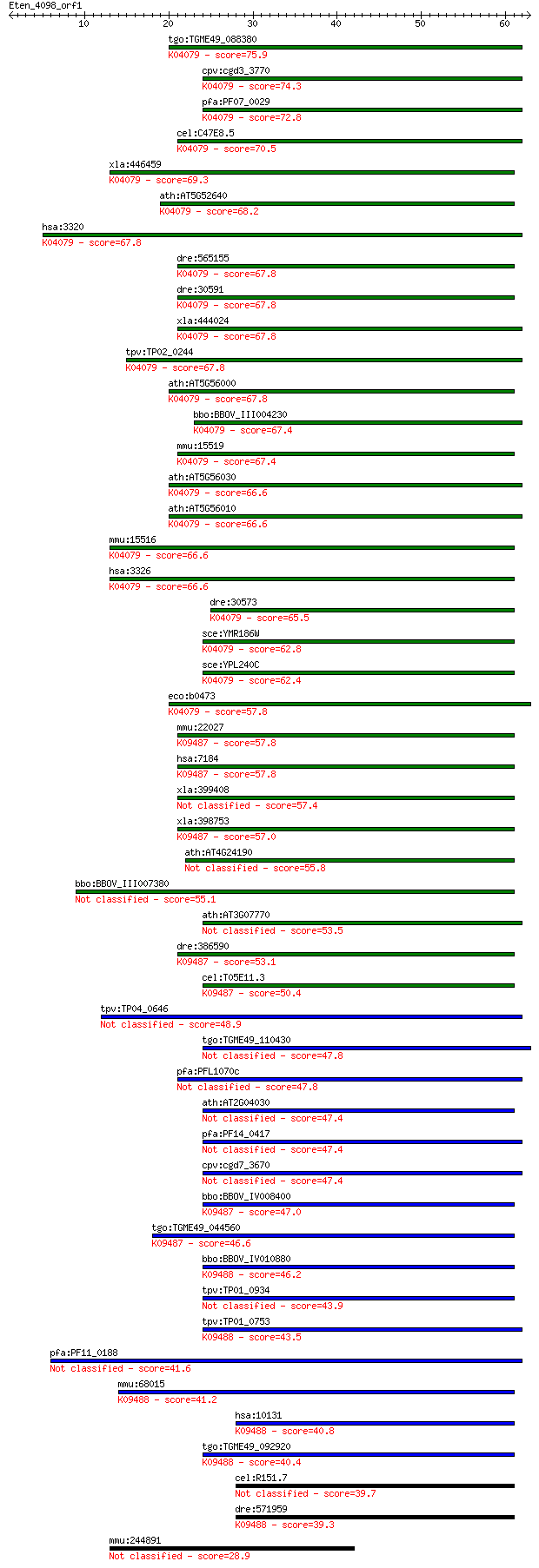
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular cha... 75.9 3e-14
cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG 74.3 9e-14
pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperon... 72.8 3e-13
cel:C47E8.5 daf-21; abnormal DAuer Formation family member (da... 70.5 1e-12
xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90k... 69.3 3e-12
ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding... 68.2 6e-12
hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N, HS... 67.8 7e-12
dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alp... 67.8 8e-12
dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb1... 67.8 8e-12
xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a, ... 67.8 8e-12
tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperon... 67.8 9e-12
ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecu... 67.8 9e-12
bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular ... 67.4 9e-12
mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,... 67.4 1e-11
ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding; ... 66.6 2e-11
ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;... 66.6 2e-11
mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1, H... 66.6 2e-11
hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2... 66.6 2e-11
dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,... 65.5 4e-11
sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90 f... 62.8 3e-10
sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone HtpG 62.4 3e-10
eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 fam... 57.8 8e-09
mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, end... 57.8 9e-09
hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein ... 57.8 9e-09
xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protei... 57.4 1e-08
xla:398753 hypothetical protein MGC68448; K09487 heat shock pr... 57.0 1e-08
ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded prot... 55.8 3e-08
bbo:BBOV_III007380 17.m07646; heat shock protein 90 55.1
ath:AT3G07770 ATP binding 53.5 1e-07
dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61... 53.1 2e-07
cel:T05E11.3 hypothetical protein; K09487 heat shock protein 9... 50.4 1e-06
tpv:TP04_0646 heat shock protein 90 48.9
tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3 ... 47.8 8e-06
pfa:PFL1070c endoplasmin homolog precursor, putative 47.8 9e-06
ath:AT2G04030 CR88; CR88; ATP binding 47.4 1e-05
pfa:PF14_0417 HSP90 47.4 1e-05
cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide pl... 47.4 1e-05
bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 hea... 47.0 1e-05
tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3... 46.6 2e-05
bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF... 46.2 2e-05
tpv:TP01_0934 heat shock protein 90 43.9
tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-assoc... 43.5 2e-04
pfa:PF11_0188 heat shock protein 90, putative 41.6 6e-04
mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated... 41.2 7e-04
hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protei... 40.8 0.001
tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF... 40.4 0.001
cel:R151.7 hypothetical protein 39.7 0.002
dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated... 39.3 0.004
mmu:244891 Scaper, C430017I08, D530014O03Rik, Zfp291; S phase ... 28.9 3.9
> tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular chaperone
HtpG
Length=708
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/42 (85%), Positives = 38/42 (90%), Gaps = 0/42 (0%)
Query 20 MENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
M + ETFAFN+DIQQLMSLIINTFYSNKEIFLRE ISNASD
Sbjct 1 MADTETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDA 42
> cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG
Length=711
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 35/38 (92%), Positives = 36/38 (94%), Gaps = 0/38 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
ETFAFN+DIQQLMSLIINTFYSNKEIFLRE ISNASD
Sbjct 15 ETFAFNADIQQLMSLIINTFYSNKEIFLRELISNASDA 52
> pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperone
HtpG
Length=745
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 34/38 (89%), Positives = 36/38 (94%), Gaps = 0/38 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
ETFAFN+DI+QLMSLIINTFYSNKEIFLRE ISNASD
Sbjct 4 ETFAFNADIRQLMSLIINTFYSNKEIFLRELISNASDA 41
> cel:C47E8.5 daf-21; abnormal DAuer Formation family member (daf-21);
K04079 molecular chaperone HtpG
Length=702
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 33/41 (80%), Positives = 36/41 (87%), Gaps = 0/41 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
EN ETFAF ++I QLMSLIINTFYSNKEI+LRE ISNASD
Sbjct 3 ENAETFAFQAEIAQLMSLIINTFYSNKEIYLRELISNASDA 43
> xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90kDa
alpha (cytosolic), class B member 1; K04079 molecular chaperone
HtpG
Length=722
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 34/48 (70%), Positives = 38/48 (79%), Gaps = 0/48 (0%)
Query 13 PKTSVAKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
P+ + E ETFAF ++I QLMSLIINTFYSNKEIFLRE ISNASD
Sbjct 2 PEVAHNGEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASD 49
> ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding
/ unfolded protein binding; K04079 molecular chaperone HtpG
Length=705
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 31/42 (73%), Positives = 37/42 (88%), Gaps = 0/42 (0%)
Query 19 KMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
+M + ETFAF ++I QL+SLIINTFYSNKEIFLRE ISN+SD
Sbjct 5 QMADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSD 46
> hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N,
HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2; heat
shock protein 90kDa alpha (cytosolic), class A member 1;
K04079 molecular chaperone HtpG
Length=854
Score = 67.8 bits (164), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 35/61 (57%), Positives = 42/61 (68%), Gaps = 4/61 (6%)
Query 5 LCIWLCLPPKTSVAKM----ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
+ + L +P +T E ETFAF ++I QLMSLIINTFYSNKEIFLRE ISN+SD
Sbjct 117 ILLRLLMPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSD 176
Query 61 C 61
Sbjct 177 A 177
> dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alpha
2; K04079 molecular chaperone HtpG
Length=734
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 32/40 (80%), Positives = 35/40 (87%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E ETFAF ++I QLMSLIINTFYSNKEIFLRE ISN+SD
Sbjct 13 EEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSD 52
> dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb17b01,
zgc:86652; heat shock protein 90-alpha 1; K04079 molecular
chaperone HtpG
Length=726
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 32/40 (80%), Positives = 35/40 (87%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E ETFAF ++I QLMSLIINTFYSNKEIFLRE ISN+SD
Sbjct 13 EEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSD 52
> xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a,
hspc1, hspca, hspn, lap2; heat shock protein 90kDa alpha (cytosolic),
class A member 1, gene 1; K04079 molecular chaperone
HtpG
Length=729
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 32/41 (78%), Positives = 36/41 (87%), Gaps = 0/41 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
E+ ETFAF ++I QLMSLIINTFYSNKEIFLRE ISN+SD
Sbjct 16 EDVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDA 56
> tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperone
HtpG
Length=721
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 32/47 (68%), Positives = 38/47 (80%), Gaps = 0/47 (0%)
Query 15 TSVAKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
TS + ++E +AFN+DI QL+SLIIN FYSNKEIFLRE ISNASD
Sbjct 2 TSKDETPDQEVYAFNADISQLLSLIINAFYSNKEIFLRELISNASDA 48
> ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecular
chaperone HtpG
Length=699
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 31/41 (75%), Positives = 36/41 (87%), Gaps = 0/41 (0%)
Query 20 MENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
M + ETFAF ++I QL+SLIINTFYSNKEIFLRE ISN+SD
Sbjct 1 MADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSD 41
> bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular
chaperone HtpG
Length=712
Score = 67.4 bits (163), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 31/39 (79%), Positives = 35/39 (89%), Gaps = 0/39 (0%)
Query 23 KETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
+ET+AFN+DI QL+SLIIN FYSNKEIFLRE ISNASD
Sbjct 6 QETYAFNADISQLLSLIINAFYSNKEIFLRELISNASDA 44
> mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,
Hsp89, Hsp90, Hspca, hsp4; heat shock protein 90, alpha (cytosolic),
class A member 1; K04079 molecular chaperone HtpG
Length=733
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 32/40 (80%), Positives = 35/40 (87%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E ETFAF ++I QLMSLIINTFYSNKEIFLRE ISN+SD
Sbjct 15 EEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSD 54
> ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding;
K04079 molecular chaperone HtpG
Length=699
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/42 (73%), Positives = 36/42 (85%), Gaps = 0/42 (0%)
Query 20 MENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
M + ETFAF ++I QL+SLIINTFYSNKEIFLRE ISN+SD
Sbjct 1 MADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSDA 42
> ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;
K04079 molecular chaperone HtpG
Length=699
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/42 (73%), Positives = 36/42 (85%), Gaps = 0/42 (0%)
Query 20 MENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
M + ETFAF ++I QL+SLIINTFYSNKEIFLRE ISN+SD
Sbjct 1 MADAETFAFQAEINQLLSLIINTFYSNKEIFLRELISNSSDA 42
> mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1,
Hsp90, Hspcb, MGC115780; heat shock protein 90 alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 34/48 (70%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 13 PKTSVAKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
P+ E ETFAF ++I QLMSLIINTFYSNKEIFLRE ISNASD
Sbjct 2 PEEVHHGEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASD 49
> hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2,
HSPCB; heat shock protein 90kDa alpha (cytosolic), class
B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 34/48 (70%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 13 PKTSVAKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
P+ E ETFAF ++I QLMSLIINTFYSNKEIFLRE ISNASD
Sbjct 2 PEEVHHGEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNASD 49
> dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,
wu:fd59e11, wu:gcd22h07; heat shock protein 90, alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=725
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 30/36 (83%), Positives = 33/36 (91%), Gaps = 0/36 (0%)
Query 25 TFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
TFAF ++I QLMSLIINTFYSNKEIFLRE +SNASD
Sbjct 13 TFAFQAEIAQLMSLIINTFYSNKEIFLRELVSNASD 48
> sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90
family, redundant in function and nearly identical with Hsp82p,
and together they are essential; expressed constitutively
at 10-fold higher basal levels than HSP82 and induced 2-3
fold by heat shock; K04079 molecular chaperone HtpG
Length=705
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 30/37 (81%), Positives = 32/37 (86%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
ETF F ++I QLMSLIINT YSNKEIFLRE ISNASD
Sbjct 4 ETFEFQAEITQLMSLIINTVYSNKEIFLRELISNASD 40
> sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone
HtpG
Length=709
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 30/37 (81%), Positives = 32/37 (86%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
ETF F ++I QLMSLIINT YSNKEIFLRE ISNASD
Sbjct 4 ETFEFQAEITQLMSLIINTVYSNKEIFLRELISNASD 40
> eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 family;
K04079 molecular chaperone HtpG
Length=624
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 25/43 (58%), Positives = 35/43 (81%), Gaps = 0/43 (0%)
Query 20 MENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDCS 62
M+ +ET F S+++QL+ L+I++ YSNKEIFLRE ISNASD +
Sbjct 1 MKGQETRGFQSEVKQLLHLMIHSLYSNKEIFLRELISNASDAA 43
> mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, endoplasmin,
gp96; heat shock protein 90, beta (Grp94), member
1; K09487 heat shock protein 90kDa beta
Length=802
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 25/40 (62%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E E FAF +++ ++M LIIN+ Y NKEIFLRE ISNASD
Sbjct 71 EKSEKFAFQAEVNRMMKLIINSLYKNKEIFLRELISNASD 110
> hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein
90kDa beta (Grp94), member 1; K09487 heat shock protein 90kDa
beta
Length=803
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 25/40 (62%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E E FAF +++ ++M LIIN+ Y NKEIFLRE ISNASD
Sbjct 71 EKSEKFAFQAEVNRMMKLIINSLYKNKEIFLRELISNASD 110
> xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protein
90kDa beta (Grp94), member 1
Length=804
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/40 (62%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E E FAF +++ ++M LIIN+ Y NKEIFLRE ISNASD
Sbjct 71 EKSEKFAFQAEVNRMMKLIINSLYKNKEIFLRELISNASD 110
> xla:398753 hypothetical protein MGC68448; K09487 heat shock
protein 90kDa beta
Length=805
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/40 (62%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E E FAF +++ ++M LIIN+ Y NKEIFLRE ISNASD
Sbjct 71 EKSEKFAFQAEVNRMMKLIINSLYKNKEIFLRELISNASD 110
> ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded protein
binding
Length=823
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 24/39 (61%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 22 NKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
N E F F +++ +LM +IIN+ YSNK+IFLRE ISNASD
Sbjct 75 NAEKFEFQAEVSRLMDIIINSLYSNKDIFLRELISNASD 113
> bbo:BBOV_III007380 17.m07646; heat shock protein 90
Length=795
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 21/52 (40%), Positives = 39/52 (75%), Gaps = 0/52 (0%)
Query 9 LCLPPKTSVAKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
L L + K+EN++T+ F +++ ++M +I+N+ Y++K+IFLRE +SNA+D
Sbjct 106 LNLDSPSPCVKVENEQTYPFQAEVSRVMDIIVNSLYTDKDIFLRELVSNAAD 157
> ath:AT3G07770 ATP binding
Length=799
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/38 (57%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
E F + +++ +LM LI+N+ YSNKE+FLRE ISNASD
Sbjct 95 EKFEYQAEVSRLMDLIVNSLYSNKEVFLRELISNASDA 132
> dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61d09,
wu:fq25g01; heat shock protein 90, beta (grp94), member
1; K09487 heat shock protein 90kDa beta
Length=793
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 23/40 (57%), Positives = 30/40 (75%), Gaps = 0/40 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
+ E AF +++ ++M LIIN+ Y NKEIFLRE ISNASD
Sbjct 71 DKAEKHAFQAEVNRMMKLIINSLYKNKEIFLRELISNASD 110
> cel:T05E11.3 hypothetical protein; K09487 heat shock protein
90kDa beta
Length=760
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 22/37 (59%), Positives = 28/37 (75%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E F +++ ++M LIIN+ Y NKEIFLRE ISNASD
Sbjct 62 EKHEFQAEVNRMMKLIINSLYRNKEIFLRELISNASD 98
> tpv:TP04_0646 heat shock protein 90
Length=913
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 38/50 (76%), Gaps = 3/50 (6%)
Query 12 PPKTSVAKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
PP+ S++ ++T+ F +++ ++M +I+N+ Y++++IFLRE +SN++D
Sbjct 122 PPEVSLS---GEQTYPFQAEVSRVMDIIVNSLYTDRDIFLRELVSNSADA 168
> tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3
2.7.13.3)
Length=1100
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 32/39 (82%), Gaps = 0/39 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDCS 62
E F F +++++++ +I+N+ Y+++++FLRE ISNA+D S
Sbjct 246 EIFPFQAEVKRVLDIIVNSLYTDRDVFLRELISNAADAS 284
> pfa:PFL1070c endoplasmin homolog precursor, putative
Length=821
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 31/41 (75%), Gaps = 0/41 (0%)
Query 21 ENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
E+ E+ + +++ +LM +I+N+ Y+ KE+FLRE ISNA+D
Sbjct 70 ESMESHQYQTEVTRLMDIIVNSLYTQKEVFLRELISNAADA 110
> ath:AT2G04030 CR88; CR88; ATP binding
Length=780
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 18/37 (48%), Positives = 30/37 (81%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E F + +++ +L+ LI+++ YS+KE+FLRE +SNASD
Sbjct 77 EKFEYQAEVSRLLDLIVHSLYSHKEVFLRELVSNASD 113
> pfa:PF14_0417 HSP90
Length=927
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
E + F +++ ++M +I+N+ Y++K++FLRE ISNASD
Sbjct 100 EKYNFKAEVNKVMDIIVNSLYTDKDVFLRELISNASDA 137
> cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide
plus ER retention motif
Length=787
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 30/38 (78%), Gaps = 0/38 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
E++ F +++ +LM +IIN+ YS K++FLRE +SN++D
Sbjct 87 ESYEFQTEVSRLMDIIINSLYSQKDVFLRELLSNSADA 124
> bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 heat
shock protein 90kDa beta
Length=795
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 29/37 (78%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E+ + +D ++M +I+N+ YSNK++FLRE ISN++D
Sbjct 89 ESHTYQADFARVMDIIVNSLYSNKDVFLRELISNSAD 125
> tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3);
K09487 heat shock protein 90kDa beta
Length=847
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 32/43 (74%), Gaps = 0/43 (0%)
Query 18 AKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
A +++E+ + +++ +LM +IIN+ Y+ +E+FLRE ISNA D
Sbjct 80 AVQKSQESHQYQTEVSRLMDIIINSLYTQREVFLRELISNAVD 122
> bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF
receptor-associated protein 1
Length=623
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 31/37 (83%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
+T+ F ++IQ+L+ ++ ++ Y++KE+F+RE ISNASD
Sbjct 16 DTYEFKAEIQKLLQIVAHSLYTDKEVFVRELISNASD 52
> tpv:TP01_0934 heat shock protein 90
Length=1009
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 30/37 (81%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E + + +++ +L+ +I+N+ YS+K+IFLRE +SN++D
Sbjct 81 EKYEYQAEVTRLLDIIVNSLYSSKDIFLRELVSNSAD 117
> tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-associated
protein 1
Length=724
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASDC 61
+ + F ++ Q+L+ ++ ++ Y++KE+F+RE ISNASD
Sbjct 77 DVYQFKAETQKLLQIVAHSLYTDKEVFVRELISNASDA 114
> pfa:PF11_0188 heat shock protein 90, putative
Length=930
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 40/70 (57%), Gaps = 14/70 (20%)
Query 6 CIWLCLPPKTSV--------AKMENK------ETFAFNSDIQQLMSLIINTFYSNKEIFL 51
C +CL K +V +KM + E + F ++ ++L+ ++ ++ Y++KE+F+
Sbjct 39 CSIVCLRKKMNVELKKICEISKMNKRNYSSECENYEFKAETKKLLQIVAHSLYTDKEVFI 98
Query 52 REWISNASDC 61
RE ISN+SD
Sbjct 99 RELISNSSDA 108
> mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=706
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 31/47 (65%), Gaps = 6/47 (12%)
Query 14 KTSVAKMENKETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
+ SV+K E F ++ ++L+ ++ + YS KE+F+RE ISNASD
Sbjct 84 RGSVSKHE------FQAETKKLLDIVARSLYSEKEVFIRELISNASD 124
> hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protein
1; K09488 TNF receptor-associated protein 1
Length=704
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 28 FNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
F ++ ++L+ ++ + YS KE+F+RE ISNASD
Sbjct 90 FQAETKKLLDIVARSLYSEKEVFIRELISNASD 122
> tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF
receptor-associated protein 1
Length=861
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 28/37 (75%), Gaps = 0/37 (0%)
Query 24 ETFAFNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
E F ++ ++L+ ++ ++ Y++KE+F+RE ISNA+D
Sbjct 159 EVHTFKAETKKLLHIVTHSLYTDKEVFVRELISNAAD 195
> cel:R151.7 hypothetical protein
Length=479
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 28 FNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
F ++ + LM ++ + YS+ E+F+RE ISNASD
Sbjct 47 FQAETRNLMDIVAKSLYSHSEVFVRELISNASD 79
> dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=719
Score = 39.3 bits (90), Expect = 0.004, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 28 FNSDIQQLMSLIINTFYSNKEIFLREWISNASD 60
F ++ ++L+ ++ + YS KE+F+RE ISN SD
Sbjct 105 FQAETKKLLDIVARSLYSEKEVFIRELISNGSD 137
> mmu:244891 Scaper, C430017I08, D530014O03Rik, Zfp291; S phase
cyclin A-associated protein in the ER
Length=1398
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 2/31 (6%)
Query 13 PKTSVAKMENKETF--AFNSDIQQLMSLIIN 41
PK S +M+NK + AFNS +Q L+S ++N
Sbjct 1077 PKISTQEMKNKPSQGDAFNSRVQDLISYVVN 1107
Lambda K H
0.322 0.132 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2060478756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40