bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4068_orf3
Length=194
Score E
Sequences producing significant alignments: (Bits) Value
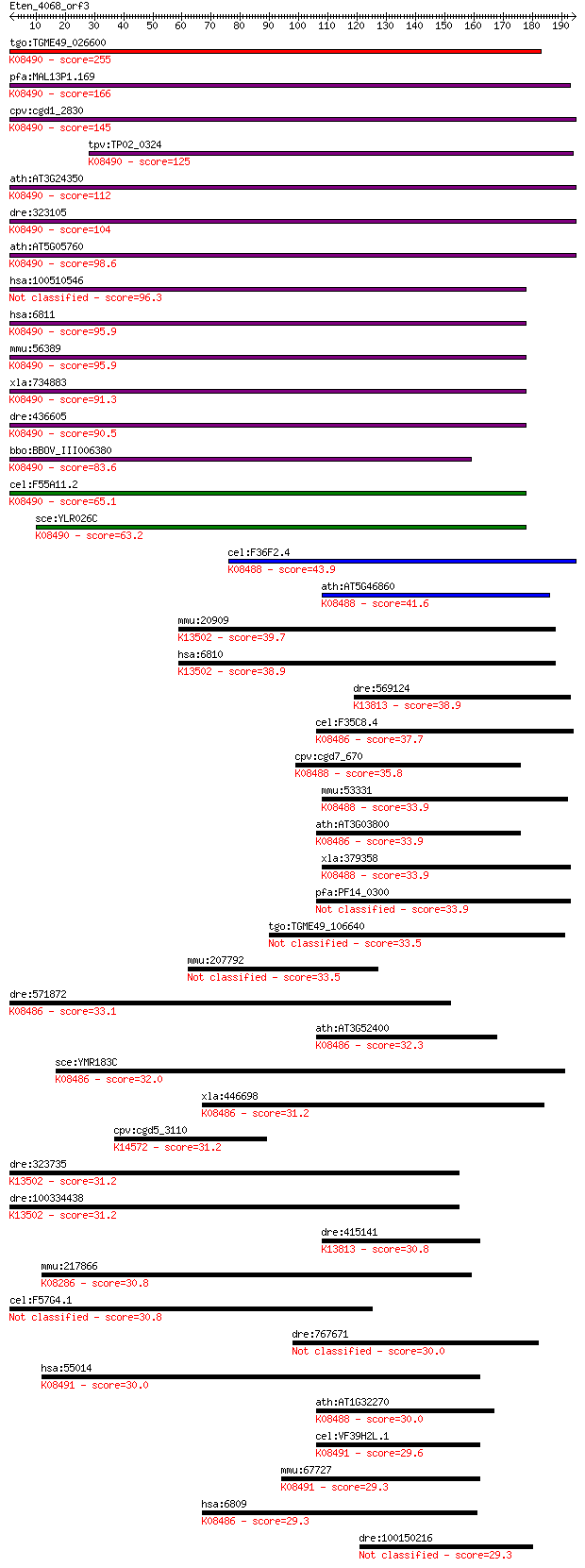
tgo:TGME49_026600 syntaxin, putative ; K08490 syntaxin 5 255 8e-68
pfa:MAL13P1.169 PfSyn5, PfStx5; Qa-SNARE protein, putative; K0... 166 4e-41
cpv:cgd1_2830 syntaxin 5A ortholog, possible transmembrane dom... 145 7e-35
tpv:TP02_0324 syntaxin 5; K08490 syntaxin 5 125 7e-29
ath:AT3G24350 SYP32; SYP32 (SYNTAXIN OF PLANTS 32); SNAP recep... 112 1e-24
dre:323105 stx5al, wu:fb81g12, zgc:73276; syntaxin 5A, like; K... 104 2e-22
ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP recep... 98.6 1e-20
hsa:100510546 syntaxin-5-like 96.3 6e-20
hsa:6811 STX5, SED5, STX5A; syntaxin 5; K08490 syntaxin 5 95.9
mmu:56389 Stx5a, 0610031F24Rik, D19Ertd627e, Stx5; syntaxin 5A... 95.9 8e-20
xla:734883 stx5, MGC114979; syntaxin 5; K08490 syntaxin 5 91.3
dre:436605 stx5a, wu:fj81f08, zgc:91864; syntaxin 5A; K08490 s... 90.5 3e-18
bbo:BBOV_III006380 17.m07568; hypothetical protein; K08490 syn... 83.6 4e-16
cel:F55A11.2 syn-3; SYNtaxin family member (syn-3); K08490 syn... 65.1 1e-10
sce:YLR026C SED5; Sed5p; K08490 syntaxin 5 63.2
cel:F36F2.4 syn-13; SYNtaxin family member (syn-13); K08488 sy... 43.9 3e-04
ath:AT5G46860 VAM3; VAM3; SNAP receptor; K08488 syntaxin 7 41.6
mmu:20909 Stx4a, Stx4, Syn-4, Syn4; syntaxin 4A (placental); K... 39.7 0.006
hsa:6810 STX4, STX4A, p35-2; syntaxin 4; K13502 syntaxin 4 38.9
dre:569124 Syntaxin-12-like; K13813 syntaxin 12/13 38.9
cel:F35C8.4 syn-1; SYNtaxin family member (syn-1); K08486 synt... 37.7 0.023
cpv:cgd7_670 t-SNARE domain followed by hydrophobic stretch ; ... 35.8 0.091
mmu:53331 Stx7, AI315064, AI317144, AW107239, Syn7; syntaxin 7... 33.9 0.32
ath:AT3G03800 SYP131; SYP131 (SYNTAXIN OF PLANTS 131); SNAP re... 33.9 0.33
xla:379358 stx7, MGC53161; syntaxin 7; K08488 syntaxin 7 33.9
pfa:PF14_0300 syn11, stx4; Qa-SNARE protein, putative 33.9 0.34
tgo:TGME49_106640 vesicle transport v-SNARE domain-containing ... 33.5 0.41
mmu:207792 6820428L09, mKIAA1614; cDNA sequence BC034090 33.5 0.42
dre:571872 zgc:165520; K08486 syntaxin 1B/2/3 33.1 0.58
ath:AT3G52400 SYP122; SYP122 (SYNTAXIN OF PLANTS 122); SNAP re... 32.3 1.0
sce:YMR183C SSO2; Plasma membrane t-SNARE involved in fusion o... 32.0 1.3
xla:446698 stx2, MGC83561, stx1b2; syntaxin 2; K08486 syntaxin... 31.2 2.3
cpv:cgd5_3110 MDN1, midasin ; K14572 midasin 31.2 2.5
dre:323735 MGC56272, STX4A, fc08c10, si:dkey-220f10.3, wu:fc08... 31.2 2.6
dre:100334438 syntaxin 4A (placental)-like; K13502 syntaxin 4 31.2
dre:415141 stx12, zgc:86632; syntaxin 12; K13813 syntaxin 12/13 30.8
mmu:217866 Cdc42bpb, MRCKb; CDC42 binding protein kinase beta ... 30.8 3.3
cel:F57G4.1 hypothetical protein 30.8 3.4
dre:767671 ccdc146, MGC152979, si:ch211-276e22.2, zgc:152979; ... 30.0 4.6
hsa:55014 STX17, FLJ20651, MGC102796, MGC126613, MGC126615; sy... 30.0 5.7
ath:AT1G32270 ATSYP24; SNAP receptor/ protein binding; K08488 ... 30.0 5.8
cel:VF39H2L.1 hypothetical protein; K08491 syntaxin 17 29.6
mmu:67727 Stx17, 4833418L03Rik, 6330411F21Rik, 9030425C21Rik, ... 29.3 8.3
hsa:6809 STX3, FLJ30906, STX3A; syntaxin 3; K08486 syntaxin 1B... 29.3 8.5
dre:100150216 hypothetical LOC100150216 29.3 9.4
> tgo:TGME49_026600 syntaxin, putative ; K08490 syntaxin 5
Length=283
Score = 255 bits (651), Expect = 8e-68, Method: Compositional matrix adjust.
Identities = 122/188 (64%), Positives = 158/188 (84%), Gaps = 6/188 (3%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGVSGQSKQHYITVTEVLKTRLLDVTKEFKDILMLRTEN 60
I+K+I LNCK+D LE+ A +G GQS+QHY T+ ++LK RLLDVTKEFKD+L+LRTEN
Sbjct 84 IKKSITELNCKIDYLEQIAKDSGSEGQSRQHYNTMVDMLKGRLLDVTKEFKDVLLLRTEN 143
Query 61 LKRQDKRRNLYSFGGD-----SGTAEAEEMYDIEGGQRTTLVS-RPATAYSQARAEAVEN 114
+K+QD+RRNLYSF G S ++ YD+EGG++T LV+ R +++Y+Q+RAEAVEN
Sbjct 144 MKKQDERRNLYSFAGSLNPSSSAYGKSSGDYDLEGGEKTQLVAQRDSSSYAQSRAEAVEN 203
Query 115 VQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNRISSNRSL 174
VQRVIGELA+IFQRVATMI+HQ+EMIQRI QDIDTSM++IR+GQ ELLN++NRISSNR+L
Sbjct 204 VQRVIGELATIFQRVATMISHQDEMIQRIDQDIDTSMHNIRQGQTELLNYFNRISSNRAL 263
Query 175 IIKVFLLL 182
I+KVF +L
Sbjct 264 ILKVFAIL 271
> pfa:MAL13P1.169 PfSyn5, PfStx5; Qa-SNARE protein, putative;
K08490 syntaxin 5
Length=281
Score = 166 bits (421), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 82/200 (41%), Positives = 129/200 (64%), Gaps = 8/200 (4%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGVSG-QSKQHYITVTEVLKTRLLDVTKEFKDILMLRTE 59
+++TI + ++D L + + +S QSK H + LK RL D TK+FKD+L +R+E
Sbjct 81 VKQTITDVTNELDLLVQYSCNLNISNPQSKTHIDNIISSLKNRLFDFTKKFKDVLQIRSE 140
Query 60 NLKRQDKRRNLYSFGGDSGTAEAEEM-------YDIEGGQRTTLVSRPATAYSQARAEAV 112
++K+Q RR +YS+ + T + DIE GQ+ L +Y +RA+A+
Sbjct 141 HIKKQVNRRKMYSYTSNEATFNNDNYKFTPLGDIDIESGQQQVLKQPSKHSYLHSRADAM 200
Query 113 ENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNRISSNR 172
EN+Q+VIG+LA +FQ+VATM+ Q+EMI+RI +DID S+ + REGQN LL ++NR++S R
Sbjct 201 ENIQKVIGDLAQMFQKVATMVTQQDEMIKRIDEDIDISLTNTREGQNYLLTYFNRLTSTR 260
Query 173 SLIIKVFLLLMAFILFFVFF 192
+LI+++F + I+FFV F
Sbjct 261 TLILQIFACIFILIVFFVIF 280
> cpv:cgd1_2830 syntaxin 5A ortholog, possible transmembrane domain
or GPI at C-terminus ; K08490 syntaxin 5
Length=329
Score = 145 bits (367), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 99/236 (41%), Positives = 141/236 (59%), Gaps = 42/236 (17%)
Query 1 IRKTINTLNCKVDTLEKAA----AAAGVSGQSKQHYITVTEVLKTRLLDVTKEFKDILML 56
I+ ++ LN +++ L++ A ++G QS QHY+T+ E LK R+LD+T++FKD L
Sbjct 94 IKTSVTELNSRLEVLQQHAQQGFPSSGGYYQSSQHYMTMVETLKARMLDITRDFKDTLQK 153
Query 57 RTENLKRQDKRRNLY----------SFG---GDSGTAEAEEM---------YDIEGG--- 91
RTE +++QD RRNLY FG GDS T+ + +DIE G
Sbjct 154 RTEVIQQQDWRRNLYSYSSNSNNLSGFGSSIGDSKTSFSSGTKYSKMSRVPFDIESGEHG 213
Query 92 -------------QRTTLVSRPATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEE 138
Q + +Y+++RAEAVENVQR+IGELA IFQ+VA M+ QEE
Sbjct 214 MDQGGTEFEFGAAQSMMQHQNQSFSYARSRAEAVENVQRMIGELAQIFQKVAGMVTQQEE 273
Query 139 MIQRIGQDIDTSMYHIREGQNELLNFYNRISSNRSLIIKVFLLLMAFILFFVFFLS 194
MIQRI +DI + ++ G EL+ +YN + SNR LIIK+FLLL+AFI+F+V FL+
Sbjct 274 MIQRIDEDISNTFSNVEHGHAELIKYYNYVKSNRGLIIKLFLLLIAFIIFYVIFLT 329
> tpv:TP02_0324 syntaxin 5; K08490 syntaxin 5
Length=285
Score = 125 bits (315), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 68/181 (37%), Positives = 106/181 (58%), Gaps = 19/181 (10%)
Query 28 SKQHYITVTEVLKTRLLDVTKEFKDILMLRTENLKRQDKRRNLYSFGG-DSGTA------ 80
SK+HY ++ L+ + +VT FK+ L R + ++ Q+ RR LY+ D+ T+
Sbjct 108 SKKHYTSIIFQLRNDIFNVTNTFKETLHQRAQIMQEQENRRKLYAINDMDAQTSGIGRKR 167
Query 81 -------EAEEMYDIEGGQRTTLVSRPATAY-SQARAEAVENVQRVIGELASIFQRVATM 132
EAE+ D+E G + + P+T+ S AR EA+ NVQR IG+L+ IFQ+V
Sbjct 168 FMLQQDLEAEQQLDLESG----ITAAPSTSVISNAREEAIANVQRAIGDLSQIFQKVTAY 223
Query 133 IAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNRISSNRSLIIKVFLLLMAFILFFVFF 192
+ Q+EMI RI D + S+ ++R +NELL +Y RISSNR LIIK+ +L+ ++ F
Sbjct 224 VTQQDEMINRIDFDTEVSLSNVRSAKNELLKYYRRISSNRGLIIKILILVTVLTCLYIMF 283
Query 193 L 193
+
Sbjct 284 V 284
> ath:AT3G24350 SYP32; SYP32 (SYNTAXIN OF PLANTS 32); SNAP receptor;
K08490 syntaxin 5
Length=347
Score = 112 bits (279), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 72/245 (29%), Positives = 124/245 (50%), Gaps = 51/245 (20%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGVSGQSKQ------HYITVTEVLKTRLLDVTKEFKDIL 54
I++ I+ LN + L+ ++ G + + H TV + LK RL+D TKEFKD+L
Sbjct 103 IKQEISALNSALVDLQLFRSSQNDEGNNSRDRDKSTHSATVVDDLKYRLMDTTKEFKDVL 162
Query 55 MLRTENLKRQDKRRNLYS----------------FGGDSGTAEAEEMYDIEG-------- 90
+RTEN+K + RR L+S + +E+ + G
Sbjct 163 TMRTENMKVHESRRQLFSSNASKESTNPFVRQRPLAAKAAASESVPLPWANGSSSSSSQL 222
Query 91 -------GQRTTLVSRPAT--------------AYSQARAEAVENVQRVIGELASIFQRV 129
G+ + L+ + Y Q RAEA+ V+ I EL+SIF ++
Sbjct 223 VPWKPGEGESSPLLQQSQQQQQQQQQQMVPLQDTYMQGRAEALHTVESTIHELSSIFTQL 282
Query 130 ATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNRISSNRSLIIKVFLLLMAFILFF 189
ATM++ Q E+ RI Q+++ ++ ++ Q++L + N ISSNR L++K+F +L+AF++ F
Sbjct 283 ATMVSQQGEIAIRIDQNMEDTLANVEGAQSQLARYLNSISSNRWLMMKIFFVLIAFLMIF 342
Query 190 VFFLS 194
+FF++
Sbjct 343 LFFVA 347
> dre:323105 stx5al, wu:fb81g12, zgc:73276; syntaxin 5A, like;
K08490 syntaxin 5
Length=298
Score = 104 bits (259), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 63/215 (29%), Positives = 119/215 (55%), Gaps = 22/215 (10%)
Query 1 IRKTINTLNCKVDTLEKAAAA-AGVSGQSKQ-HYITVTEVLKTRLLDVTKEFKDILMLRT 58
+++ IN+LN ++ L++ + + +G+ Q H T+ L+++L ++ +FK +L +RT
Sbjct 85 VKQDINSLNKQIAGLQELVRSRSAQNGRHLQTHSNTIVVSLQSKLASMSSDFKSVLEVRT 144
Query 59 ENLKRQDKRRNLYS-------------------FGGDSGTAEAEEMYDIEGGQRTTLVSR 99
ENLK+Q R+ +S DS + D+ Q+ LV+
Sbjct 145 ENLKQQRSRQEQFSQTPASASAFHTNSFNNSVLMQDDSKKTDISIDMDLNSSQQMQLVN- 203
Query 100 PATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQN 159
+Y Q RA+ ++N++ I EL SIFQ++A M+ QEE + RI +++ + ++
Sbjct 204 ERDSYIQNRADTMQNIESTIVELGSIFQQLAHMVKEQEETVHRIDANVEDTQLNVDLAHT 263
Query 160 ELLNFYNRISSNRSLIIKVFLLLMAFILFFVFFLS 194
E+L ++ +S+NR L+IK+FL+L+ F + FV F++
Sbjct 264 EILKYFQSVSNNRWLLIKMFLVLVIFFIVFVLFMT 298
> ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP receptor;
K08490 syntaxin 5
Length=336
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 71/245 (28%), Positives = 119/245 (48%), Gaps = 51/245 (20%)
Query 1 IRKTINTLN---CKVDTLEKAAAAAGVSGQSK-QHYITVTEVLKTRLLDVTKEFKDILML 56
IR I LN + TL+ A G Q + HY V + LKTRL+ TK+ +D+L
Sbjct 92 IRNDITGLNMALSDLQTLQNMELADGNYSQDQVGHYTAVCDDLKTRLMGATKQLQDVLTT 151
Query 57 RTENLKRQDKRRNLYSFGG--DS----------------------GTAEAEEMYDIEGG- 91
R+EN+K + R+ L+S DS G + + + G
Sbjct 152 RSENMKAHENRKQLFSTKNAVDSPPQNNAKSVPEPPPWSSSSNPFGNLQQPLLPPLNTGA 211
Query 92 -------QRTTLVSRPATA---------------YSQARAEAVENVQRVIGELASIFQRV 129
+R+ + + P+ YSQ+RA A+ +V+ I EL+ IF ++
Sbjct 212 PPGSQLRRRSAIENAPSQQMEMSLLQQTVPKQENYSQSRAVALHSVESRITELSGIFPQL 271
Query 130 ATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNRISSNRSLIIKVFLLLMAFILFF 189
ATM+ Q E+ RI ++D S+ ++ ++ LL RISSNR L++K+F +++ F++ F
Sbjct 272 ATMVTQQGELAIRIDDNMDESLVNVEGARSALLQHLTRISSNRWLMMKIFAVIILFLIVF 331
Query 190 VFFLS 194
+FF++
Sbjct 332 LFFVA 336
> hsa:100510546 syntaxin-5-like
Length=295
Score = 96.3 bits (238), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 64/201 (31%), Positives = 109/201 (54%), Gaps = 25/201 (12%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGV-SGQSKQ-HYITVTEVLKTRLLDVTKEFKDILMLRT 58
I++ IN+LN ++ L+ A G SG+ Q H T+ L+++L ++ +FK +L +RT
Sbjct 79 IKQDINSLNKQIAQLQDFVRAKGSQSGRHLQTHSNTIVVSLQSKLASMSNDFKSVLEVRT 138
Query 59 ENLKRQDKRRNLYS-------------FGGDSGTAEAE---------EMYDIEGGQRTTL 96
ENLK+Q RR +S GG + AE +M D Q+ L
Sbjct 139 ENLKQQRSRREQFSRAPVSALPLAPNHLGGGAVVLGAESHASKDVAIDMMDSRTSQQLQL 198
Query 97 VSRPATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIRE 156
+ +Y Q+RA+ ++N++ I EL SIFQ++A M+ QEE IQRI +++ + +
Sbjct 199 IDEQ-DSYIQSRADTMQNIESTIVELGSIFQQLAHMVKEQEETIQRIDENVLGAQLDVEA 257
Query 157 GQNELLNFYNRISSNRSLIIK 177
+E+L ++ ++SNR L++K
Sbjct 258 AHSEILKYFQSVTSNRWLMVK 278
> hsa:6811 STX5, SED5, STX5A; syntaxin 5; K08490 syntaxin 5
Length=355
Score = 95.9 bits (237), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 64/201 (31%), Positives = 109/201 (54%), Gaps = 25/201 (12%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGV-SGQSKQ-HYITVTEVLKTRLLDVTKEFKDILMLRT 58
I++ IN+LN ++ L+ A G SG+ Q H T+ L+++L ++ +FK +L +RT
Sbjct 139 IKQDINSLNKQIAQLQDFVRAKGSQSGRHLQTHSNTIVVSLQSKLASMSNDFKSVLEVRT 198
Query 59 ENLKRQDKRRNLYS-------------FGGDSGTAEAE---------EMYDIEGGQRTTL 96
ENLK+Q RR +S GG + AE +M D Q+ L
Sbjct 199 ENLKQQRSRREQFSRAPVSALPLAPNHLGGGAVVLGAESHASKDVAIDMMDSRTSQQLQL 258
Query 97 VSRPATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIRE 156
+ +Y Q+RA+ ++N++ I EL SIFQ++A M+ QEE IQRI +++ + +
Sbjct 259 IDE-QDSYIQSRADTMQNIESTIVELGSIFQQLAHMVKEQEETIQRIDENVLGAQLDVEA 317
Query 157 GQNELLNFYNRISSNRSLIIK 177
+E+L ++ ++SNR L++K
Sbjct 318 AHSEILKYFQSVTSNRWLMVK 338
> mmu:56389 Stx5a, 0610031F24Rik, D19Ertd627e, Stx5; syntaxin
5A; K08490 syntaxin 5
Length=355
Score = 95.9 bits (237), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 64/201 (31%), Positives = 108/201 (53%), Gaps = 25/201 (12%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGV-SGQSKQ-HYITVTEVLKTRLLDVTKEFKDILMLRT 58
I++ IN+LN ++ L+ A G SG+ Q H T+ L+++L ++ +FK +L +RT
Sbjct 139 IKQDINSLNKQIAQLQDFVRAKGSQSGRHLQTHSNTIVVSLQSKLASMSNDFKSVLEVRT 198
Query 59 ENLKRQDKRRNLYS-------------FGGDSGTAEAE---------EMYDIEGGQRTTL 96
ENLK+Q RR +S GG AE +M D Q+ L
Sbjct 199 ENLKQQRNRREQFSRAPVSALPLAPNNLGGGPIILGAESRASRDVAIDMMDPRTSQQLQL 258
Query 97 VSRPATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIRE 156
+ +Y Q+RA+ ++N++ I EL SIFQ++A M+ QEE IQRI +++ + +
Sbjct 259 IDE-QDSYIQSRADTMQNIESTIVELGSIFQQLAHMVKEQEETIQRIDENVLGAQLDVEA 317
Query 157 GQNELLNFYNRISSNRSLIIK 177
+E+L ++ ++SNR L++K
Sbjct 318 AHSEILKYFQSVTSNRWLMVK 338
> xla:734883 stx5, MGC114979; syntaxin 5; K08490 syntaxin 5
Length=298
Score = 91.3 bits (225), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 62/198 (31%), Positives = 107/198 (54%), Gaps = 22/198 (11%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGV-SGQSKQ-HYITVTEVLKTRLLDVTKEFKDILMLRT 58
I++ I +LN ++ L+ A G SG+ Q H TV L+++L ++ +FK +L +RT
Sbjct 85 IKQDIGSLNQQIAQLQSFVRARGSQSGRHLQTHSNTVVVSLQSKLASMSNDFKSVLEVRT 144
Query 59 ENLKRQDKRRNLYSFGG-----------------DSGTAEAEEMYDIEG--GQRTTLVSR 99
ENLK+Q RR +S G D + E +++ Q+ L+
Sbjct 145 ENLKQQRSRREHFSQGQVALPLHHNSLGPSVLLQDDSRRQGEVTIEMDSRVSQQLQLID- 203
Query 100 PATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQN 159
+Y Q+RA+ ++N++ I EL SIFQ++A M+ QEE IQRI +++ + ++
Sbjct 204 EQDSYIQSRADTMQNIESTIVELGSIFQQLAHMVKEQEETIQRIDGNVEDTQLNVEGAHQ 263
Query 160 ELLNFYNRISSNRSLIIK 177
E+L ++ ++SNR L+IK
Sbjct 264 EILKYFQSVTSNRWLMIK 281
> dre:436605 stx5a, wu:fj81f08, zgc:91864; syntaxin 5A; K08490
syntaxin 5
Length=302
Score = 90.5 bits (223), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 61/200 (30%), Positives = 107/200 (53%), Gaps = 23/200 (11%)
Query 1 IRKTINTLNCKVDTLEK-AAAAAGVSGQSKQ-HYITVTEVLKTRLLDVTKEFKDILMLRT 58
I++ IN+LN ++ L+ + +G +G+ Q H T+ L+++L ++ +FK +L +RT
Sbjct 86 IKQDINSLNKQIAQLQDLVRSRSGQNGRHIQTHSNTIVVSLQSKLASMSNDFKSVLEVRT 145
Query 59 ENLKRQDKRRNLYS-------------------FGGDSGTAEAEEMYDIEGGQRTTLVS- 98
ENLK+Q RR +S +S + AE D++ +
Sbjct 146 ENLKQQRSRREHFSQAPVSASPLLANNFNSSVLMQDESRSLGAEVAIDMDSRANPLQLQL 205
Query 99 -RPATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREG 157
+Y Q+RA+ ++N++ I EL SIFQ++A M+ QEE IQRI ++D + ++
Sbjct 206 IDEQDSYIQSRADTMQNIESTIVELGSIFQQLAHMVKEQEETIQRIDANVDDTELNVEMA 265
Query 158 QNELLNFYNRISSNRSLIIK 177
E+L ++ +SSNR L+IK
Sbjct 266 HGEILKYFQSVSSNRWLMIK 285
> bbo:BBOV_III006380 17.m07568; hypothetical protein; K08490 syntaxin
5
Length=256
Score = 83.6 bits (205), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 57/176 (32%), Positives = 92/176 (52%), Gaps = 22/176 (12%)
Query 1 IRKTINTLNCKVDTLE-KAAAAAGVSGQSKQHYITVTEVLKTRLLDVTKEFKDILMLRTE 59
+++ I + K+D E K + + +QHY + L+ +L ++TK KD L R +
Sbjct 81 VKEGITAASSKIDEFETKVRSIRHKNDHVRQHYENLLGTLRKQLCELTKSLKDALYQRAQ 140
Query 60 NLKRQDKRRNLYS-------FGGDSGT---------AEAEEMYDIEGGQRTTLVSRPA-T 102
+ +Q+ RR +YS S T E + D+E G +V RP+ +
Sbjct 141 VMIQQEMRRKMYSHTDADHSINATSNTRRRFTMQPSHEDVQQLDLESG----VVERPSRS 196
Query 103 AYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQ 158
+ A+AEA+ NVQR I EL+ IFQR+ TM+ Q+EMIQRI D + S+ ++ G+
Sbjct 197 VIADAKAEALANVQRAISELSQIFQRMTTMVTQQDEMIQRIDMDTEDSLANVIAGK 252
> cel:F55A11.2 syn-3; SYNtaxin family member (syn-3); K08490 syntaxin
5
Length=413
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 103/206 (50%), Gaps = 29/206 (14%)
Query 1 IRKTINTLNCKVDTLEKAAA--AAGVSGQSKQHYITVTEVLKTRLLDVTKEFKDILMLRT 58
++ I LN ++ L++ + A + Q+ H V L+++L +V K+++ +L + T
Sbjct 191 VKSDITGLNKQIGQLQEFSKRRAGNMKNQNSGHIQLVVVGLQSKLANVGKDYQSVLEIST 250
Query 59 ENLKRQDKRRNLYSFGG---------DSG----------------TAEAEEMYDIEGGQ- 92
E +K + RR+ +S G SG ++ A +M + Q
Sbjct 251 ETMKAEKNRRDKFSSGAAVPMGLPSSSSGANVRSKLLQDDEQHGSSSIALDMGALSNMQS 310
Query 93 RTTLVSRPAT-AYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSM 151
+ T+ R ++ Y+QAR+ + ++ I EL IF ++A++++ Q EMI RI +++ +
Sbjct 311 QQTMQQRDSSLEYAQARSNTMATIEGSISELGQIFSQLASLVSEQGEMITRIDSNVEDTA 370
Query 152 YHIREGQNELLNFYNRISSNRSLIIK 177
+I +EL+ + IS NR L+I+
Sbjct 371 LNIDMAHSELVRYLQNISKNRWLMIQ 396
> sce:YLR026C SED5; Sed5p; K08490 syntaxin 5
Length=340
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 58/216 (26%), Positives = 93/216 (43%), Gaps = 50/216 (23%)
Query 10 CKVDTLEKAAAAAGVSGQSK------QHYITVTEVLKTRLLDVTKEFKDILMLRTENLKR 63
++ L+K S QS QH V +L T++ +++ FKD+L R + L+
Sbjct 111 VQLSQLKKTDVNGNTSNQSSKQPSAVQHSKNVVNLLNTQMKNISGSFKDVLEER-QRLEM 169
Query 64 QDKRRNLYSFGGDSGTAEAEEM-----------YDIEGGQRTTLV--------------- 97
+K R D+G A A++ Y+ T+L+
Sbjct 170 ANKDR-WQKLTTDTGHAPADDQTQSNHAADLTTYNNSNPFMTSLLDESSEKNNNSSNQGE 228
Query 98 -SRPAT---------------AYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQ 141
S P Y Q R AVE ++ I E+ ++FQ++A+M+ Q E+IQ
Sbjct 229 LSFPQNDSQLMLMEEGQLSNNVYLQERNRAVETIESTIQEVGNLFQQLASMVQEQGEVIQ 288
Query 142 RIGQDIDTSMYHIREGQNELLNFYNRISSNRSLIIK 177
RI ++D +I Q ELL +++RI SNR L K
Sbjct 289 RIDANVDDIDLNISGAQRELLKYFDRIKSNRWLAAK 324
> cel:F36F2.4 syn-13; SYNtaxin family member (syn-13); K08488
syntaxin 7
Length=248
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/128 (21%), Positives = 62/128 (48%), Gaps = 9/128 (7%)
Query 76 DSGTAEAEEMYDIEGGQRTTLVSRPATAYSQA-------RAEAVENVQRVIGELASIFQR 128
D+ A YD+ G + TA Q R A++ ++R IG++ +IF
Sbjct 120 DAQAARDAAEYDMYGNNGRSGGQMQMTAQQQGNLQDMKERQNALQQLERDIGDVNAIFAE 179
Query 129 VATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLN--FYNRISSNRSLIIKVFLLLMAFI 186
+A ++ Q +M+ I +++ + ++ +G + +YN+ + + L++ F +++ FI
Sbjct 180 LANIVHEQGDMVDSIEANVEHAQIYVEQGAQNVQQAVYYNQKARQKKLLLLCFFVILIFI 239
Query 187 LFFVFFLS 194
+ +L+
Sbjct 240 IGLTLYLA 247
> ath:AT5G46860 VAM3; VAM3; SNAP receptor; K08488 syntaxin 7
Length=268
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 41/78 (52%), Gaps = 3/78 (3%)
Query 108 RAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNR 167
R + ++ + + IGE+ IF+ +A ++ Q MI IG ID S +G+++L+
Sbjct 181 REQGIQEIHQQIGEVNEIFKDLAVLVNDQGVMIDDIGTHIDNSRAATSQGKSQLVQAAKT 240
Query 168 ISSNRSLIIKVFLLLMAF 185
SN SL LLL+ F
Sbjct 241 QKSNSSLTC---LLLVIF 255
> mmu:20909 Stx4a, Stx4, Syn-4, Syn4; syntaxin 4A (placental);
K13502 syntaxin 4
Length=298
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 41/144 (28%), Positives = 69/144 (47%), Gaps = 21/144 (14%)
Query 59 ENLKRQDKRRNLYSFGGDSGTAEAEEMYDIEGGQ---------RTTLVSRPATAYSQARA 109
E ++RQ K N G E E+M D GQ + T V+R A AR
Sbjct 154 ERIRRQLKITN----AGMVSDEELEQMLD--SGQSEVFVSNILKDTQVTRQALNEISARH 207
Query 110 EAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNEL-LNFYNRI 168
++ ++R I EL IF +AT + Q EMI RI ++I +S ++ GQ + + N+
Sbjct 208 SEIQQLERSIRELHEIFTFLATEVEMQGEMINRIEKNILSSADYVERGQEHVKIALENQK 267
Query 169 SSNR-----SLIIKVFLLLMAFIL 187
+ + ++ + V +L++A I+
Sbjct 268 KARKKKVMIAICVSVTVLILAVII 291
> hsa:6810 STX4, STX4A, p35-2; syntaxin 4; K13502 syntaxin 4
Length=297
Score = 38.9 bits (89), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 42/144 (29%), Positives = 67/144 (46%), Gaps = 21/144 (14%)
Query 59 ENLKRQDKRRNLYSFGGDSGTAEAEEMYDIEGGQ---------RTTLVSRPATAYSQARA 109
E ++RQ K N G E E+M D GQ + T V+R A AR
Sbjct 154 ERIRRQLKITN----AGMVSDEELEQMLD--SGQSEVFVSNILKDTQVTRQALNEISARH 207
Query 110 EAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNEL---LNFYN 166
++ ++R I EL IF +AT + Q EMI RI ++I +S ++ GQ + L
Sbjct 208 SEIQQLERSIRELHDIFTFLATEVEMQGEMINRIEKNILSSADYVERGQEHVKTALENQK 267
Query 167 RISSNRSLI---IKVFLLLMAFIL 187
+ + LI + + ++L+A I+
Sbjct 268 KARKKKVLIAICVSITVVLLAVII 291
> dre:569124 Syntaxin-12-like; K13813 syntaxin 12/13
Length=267
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 41/76 (53%), Gaps = 2/76 (2%)
Query 119 IGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLN--FYNRISSNRSLII 176
I ++ IF+ +A MI Q +MI I +++++ H+ G +L + +Y R S R I+
Sbjct 188 IMDVNQIFKDLAVMIHDQGDMIDSIEANVESAEVHVERGAEQLQHAAYYQRKSRKRMCIL 247
Query 177 KVFLLLMAFILFFVFF 192
+ L L+A I + +
Sbjct 248 ALVLSLVATIFAIIIW 263
> cel:F35C8.4 syn-1; SYNtaxin family member (syn-1); K08486 syntaxin
1B/2/3
Length=306
Score = 37.7 bits (86), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 18/88 (20%), Positives = 46/88 (52%), Gaps = 0/88 (0%)
Query 106 QARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFY 165
++RA+ ++N++R +GELA +F + M+ Q EM+ I ++ + + ++ + +
Sbjct 208 KSRADELKNLERQMGELAQMFHDLHIMVVSQGEMVDSIVNSVENATEYAKQARGNVEEAR 267
Query 166 NRISSNRSLIIKVFLLLMAFILFFVFFL 193
N R + + + + + +L + F+
Sbjct 268 NLQKRARKMKVCIIIGSIIAVLILILFI 295
> cpv:cgd7_670 t-SNARE domain followed by hydrophobic stretch
; K08488 syntaxin 7
Length=311
Score = 35.8 bits (81), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 41/89 (46%), Gaps = 12/89 (13%)
Query 99 RPATAYSQARAEAVE------------NVQRVIGELASIFQRVATMIAHQEEMIQRIGQD 146
+P T Y+ + V+ ++Q + E IF+ +A+M+ Q E IQ +
Sbjct 198 KPPTLYTHQTFDMVDRQIAQETAIGLGHIQSQMYEANQIFKNLASMVNEQGETIQNLETT 257
Query 147 IDTSMYHIREGQNELLNFYNRISSNRSLI 175
ID ++Y ++ EL YN S SLI
Sbjct 258 IDNTVYTAKQAVGELRKAYNSSSYKFSLI 286
> mmu:53331 Stx7, AI315064, AI317144, AW107239, Syn7; syntaxin
7; K08488 syntaxin 7
Length=261
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 19/86 (22%), Positives = 46/86 (53%), Gaps = 3/86 (3%)
Query 108 RAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNF--Y 165
R ++ ++ I ++ IF+ + MI Q +MI I +++++ H+++ +L Y
Sbjct 171 RESSIRQLEADIMDINEIFKDLGMMIHEQGDMIDSIEANVESAEVHVQQANQQLSRAADY 230
Query 166 NRISSNRSLIIKVFLLLMAFILFFVF 191
R S ++L I +F+L++ ++ +
Sbjct 231 QR-KSRKTLCIIIFILVVGIVIICLI 255
> ath:AT3G03800 SYP131; SYP131 (SYNTAXIN OF PLANTS 131); SNAP
receptor; K08486 syntaxin 1B/2/3
Length=306
Score = 33.9 bits (76), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 38/70 (54%), Gaps = 0/70 (0%)
Query 106 QARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFY 165
Q R +AV ++++ + +L +F +A ++ Q EM+ I + +++ H++ G N+L
Sbjct 209 QERHDAVRDLEKKLLDLQQVFLDMAVLVDAQGEMLDNIENMVSSAVDHVQSGNNQLTKAV 268
Query 166 NRISSNRSLI 175
S+R +
Sbjct 269 KSQKSSRKWM 278
> xla:379358 stx7, MGC53161; syntaxin 7; K08488 syntaxin 7
Length=259
Score = 33.9 bits (76), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 20/87 (22%), Positives = 42/87 (48%), Gaps = 2/87 (2%)
Query 108 RAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNF--Y 165
R A+ ++ I + IF+ + M+ Q EMI I +++ + H+++ +L Y
Sbjct 169 RETAIRQLEEDIQGINEIFKDLGMMVHEQGEMIDSIEANVENAEVHVQQANQQLATAAEY 228
Query 166 NRISSNRSLIIKVFLLLMAFILFFVFF 192
R S + II L++ A ++ + +
Sbjct 229 QRKSRRKICIIIAVLVVAATVIGLIIW 255
> pfa:PF14_0300 syn11, stx4; Qa-SNARE protein, putative
Length=442
Score = 33.9 bits (76), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 50/94 (53%), Gaps = 11/94 (11%)
Query 106 QARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNF- 164
+ + ++N++ I L ++ +A +I ++ +I +I+ +++ ++E + LN
Sbjct 348 KKKYNELKNLENNILSLNELYIELAYVIKKRKNLIN----NIENNVFQVKEYTQDALNNI 403
Query 165 -----YNRISSNRSLIIKVFLLLMAFILFF-VFF 192
YN + R L +FLL++AFI+ F VFF
Sbjct 404 VDAKKYNSMIKQRILYFSIFLLIVAFIILFPVFF 437
> tgo:TGME49_106640 vesicle transport v-SNARE domain-containing
protein (EC:2.7.1.69)
Length=376
Score = 33.5 bits (75), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 54/108 (50%), Gaps = 9/108 (8%)
Query 90 GGQ--RTTLVSRPATAYSQARAEA-VENVQRVIGELASIFQRVATMIAHQEEMIQRIGQD 146
GG+ R TLV A Q + EA + + ++GE+ + ++ T + Q E +++ QD
Sbjct 269 GGEHMRQTLVQ--AGDEIQDKTEASINRTKHMVGEMEEMGAQILTKMDEQTEQLRKANQD 326
Query 147 IDTSMYHIREGQNELLNFYNRISSNR---SLIIKVFL-LLMAFILFFV 190
+D + Y++ + + + +R L + +FL L++A +L V
Sbjct 327 LDDAHYNVDRAKKTAITLAKNAAGDRFSQCLCLFIFLFLVIAIVLVCV 374
> mmu:207792 6820428L09, mKIAA1614; cDNA sequence BC034090
Length=1171
Score = 33.5 bits (75), Expect = 0.42, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 36/70 (51%), Gaps = 8/70 (11%)
Query 62 KRQDKRRNLYSFGGDSGTAEAEEMYDIEGGQRTTLVSRPATAYSQARAEAVENV-----Q 116
+R +NL+S G G + +Y +EG ++ SRPA A+ RA +VE+V
Sbjct 1011 RRATSFQNLHSLLG--GKGDRSSLYLVEGSGDSSGPSRPAKAFPH-RALSVEDVGAPSLA 1067
Query 117 RVIGELASIF 126
R +G + +F
Sbjct 1068 RTVGRVVEVF 1077
> dre:571872 zgc:165520; K08486 syntaxin 1B/2/3
Length=326
Score = 33.1 bits (74), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 41/166 (24%), Positives = 75/166 (45%), Gaps = 23/166 (13%)
Query 1 IRKTINTLNCKVDTLEKAAAA-------AGVSGQSKQHYITVTEVLKTRLLDVTKEFKDI 53
I+K N K+ ++E+ AA A + + QH I L + ++V ++ +
Sbjct 81 IKKLANNARNKLKSIEQNLAANTEERVSADMRIKKSQHAI-----LAKKFVEVMTKYNEA 135
Query 54 LMLRTENLKRQDKRRNLYSFGGDSGTAEAEEMYDIEGGQRTTL--------VSRPATAYS 105
+ E K + +R+ L G + E EEM D GG +S+ A +
Sbjct 136 QVEFREKSKGRIQRQ-LEITGKATTDEELEEMLD--GGNAAVFTAGIMDSGISKQALSEI 192
Query 106 QARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSM 151
+AR + + ++ I EL +F +A ++ +Q MI RI ++D S+
Sbjct 193 EARHKDIMRLESSIKELHDMFVDIAVLVENQGSMIDRIESNMDQSV 238
> ath:AT3G52400 SYP122; SYP122 (SYNTAXIN OF PLANTS 122); SNAP
receptor; K08486 syntaxin 1B/2/3
Length=341
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 16/64 (25%), Positives = 35/64 (54%), Gaps = 2/64 (3%)
Query 106 QARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLN-- 163
Q R +AV+++++ + EL +F +A ++ HQ + I ++ + +R G + L+
Sbjct 217 QERHDAVKDIEKSLNELHQVFLDMAVLVEHQGAQLDDIEGNVKRANSLVRSGADRLVKAR 276
Query 164 FYNR 167
FY +
Sbjct 277 FYQK 280
> sce:YMR183C SSO2; Plasma membrane t-SNARE involved in fusion
of secretory vesicles at the plasma membrane; syntaxin homolog
that is functionally redundant with Sso1p; K08486 syntaxin
1B/2/3
Length=295
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 39/187 (20%), Positives = 84/187 (44%), Gaps = 22/187 (11%)
Query 17 KAAAAAGVSGQSKQHYITVTEVLKTRLLDVTKEFKDILMLRTENLKRQDKRRNLYSFGGD 76
K A G+ +KQ E + + L + ++++ I E K Q KR+ Y+
Sbjct 103 KDAQRDGLHDSNKQ---AQAENCRQKFLKLIQDYRIIDSNYKEESKEQAKRQ--YTIIQP 157
Query 77 SGTAEAEE--MYDIEGGQRTTLV---------SRPATAYSQARAEAVENVQRVIGELASI 125
T E E + D+ G Q + ++ A A QAR + + +++ + EL +
Sbjct 158 EATDEEVEAAINDVNGQQIFSQALLNANRRGEAKTALAEVQARHQELLKLEKTMAELTQL 217
Query 126 FQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNRI--SSNRSLIIKVFLLLM 183
F + ++ Q+E + I ++++ + + +G + N+ S+ ++ K+ L++
Sbjct 218 FNDMEELVIEQQENVDVIDKNVEDAQQDVEQG----VGHTNKAVKSARKARKNKIRCLII 273
Query 184 AFILFFV 190
FI+F +
Sbjct 274 CFIIFAI 280
> xla:446698 stx2, MGC83561, stx1b2; syntaxin 2; K08486 syntaxin
1B/2/3
Length=290
Score = 31.2 bits (69), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 62/129 (48%), Gaps = 16/129 (12%)
Query 67 RRNLYSFGGDSGTAEAEEMYDIEGGQRT---------TLVSRPATAYSQARAEAVENVQR 117
+R L G + E EEM +E G + + +++ A ++R + + ++
Sbjct 150 QRQLEITGKTTTDDELEEM--LESGNPSIFTSDIISDSQITKQALNEIESRHKDIIKLES 207
Query 118 VIGELASIFQRVATMIAHQEEMIQRIGQDIDTS---MYHIREGQNELLNFYNRISSNRSL 174
I EL +F +AT++ Q EMI I ++++ + + H +E + + + ++ S R
Sbjct 208 SIRELHDMFVDIATLVESQGEMINSIEKNVENAAEYIEHAKEETKKAVKYQSK--SRRKK 265
Query 175 IIKVFLLLM 183
I FL+L+
Sbjct 266 WIAAFLVLV 274
> cpv:cgd5_3110 MDN1, midasin ; K14572 midasin
Length=2893
Score = 31.2 bits (69), Expect = 2.5, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query 37 EVLKTRLLDVTKEFKDILMLRTEN---LKRQDKRRNLYSFGGDSGTAEAEEMYDI 88
++L+ + L+ +E D L TEN L ++D+ L +FG DSG + + Y I
Sbjct 2074 QILQLKNLNRDEEIDDYFKLMTENGIQLIKEDQNYTLMTFGSDSGNSSDVKEYTI 2128
> dre:323735 MGC56272, STX4A, fc08c10, si:dkey-220f10.3, wu:fc08c10;
zgc:56272; K13502 syntaxin 4
Length=297
Score = 31.2 bits (69), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 41/165 (24%), Positives = 69/165 (41%), Gaps = 21/165 (12%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGVSGQSKQHYITVTEVLKT--RLLDVTKEFKDILMLRT 58
I+K + +L K +E+ V + QH + E L+ R + +++D
Sbjct 99 IQKKLKSLEPKKLEVEEKYIPVNVRMRRTQHGVLSREFLELMGRCNTIQAQYRD---RNV 155
Query 59 ENLKRQDKRRNLYSFGGDSGTAEAEEMYDIEGGQRTTL---------VSRPATAYSQARA 109
E +KRQ L G E E M +E GQ +R A ++R
Sbjct 156 ERIKRQ-----LKITGNSVSDDELETM--LESGQTDVFTQNILNDAKATRQALNEIESRH 208
Query 110 EAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHI 154
+ + ++R I EL +FQ +A + Q EM+ RI +I S ++
Sbjct 209 DEIIKLERSIKELHDMFQYLAMEVEAQGEMVDRIESNIKMSHDYV 253
> dre:100334438 syntaxin 4A (placental)-like; K13502 syntaxin
4
Length=297
Score = 31.2 bits (69), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 41/165 (24%), Positives = 69/165 (41%), Gaps = 21/165 (12%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGVSGQSKQHYITVTEVLKT--RLLDVTKEFKDILMLRT 58
I+K + +L K +E+ V + QH + E L+ R + +++D
Sbjct 99 IQKKLKSLEPKKLEVEEKYIPVNVRMRRTQHGVLSREFLELMGRCNTIQAQYRD---RNV 155
Query 59 ENLKRQDKRRNLYSFGGDSGTAEAEEMYDIEGGQRTTL---------VSRPATAYSQARA 109
E +KRQ L G E E M +E GQ +R A ++R
Sbjct 156 ERIKRQ-----LKITGNSVSDDELETM--LESGQTDVFTQNILNDAKATRQALNEIESRH 208
Query 110 EAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHI 154
+ + ++R I EL +FQ +A + Q EM+ RI +I S ++
Sbjct 209 DEIIKLERSIKELHDMFQYLAMEVEAQGEMVDRIESNIKMSHDYV 253
> dre:415141 stx12, zgc:86632; syntaxin 12; K13813 syntaxin 12/13
Length=266
Score = 30.8 bits (68), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 108 RAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNEL 161
R A+ ++ I ++ IF+ +A MI Q EMI I +++++ H+ G +L
Sbjct 177 RETAIRQLESDILDVNQIFKDLAVMIHDQGEMIDSIEANVESAEVHVERGAEQL 230
> mmu:217866 Cdc42bpb, MRCKb; CDC42 binding protein kinase beta
(EC:2.7.11.1); K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=1713
Score = 30.8 bits (68), Expect = 3.3, Method: Composition-based stats.
Identities = 36/163 (22%), Positives = 73/163 (44%), Gaps = 23/163 (14%)
Query 12 VDTLEKAAAAAGVSGQSKQ--HYITVTEVLKTRLLD---VTKEFKDILMLRTEN------ 60
V +L + A G S + K+ E +K+++ D + ++ +D + LR E+
Sbjct 468 VQSLHGSTRALGNSNRDKEIKRLNEELERMKSKMADSNRLERQLEDTVTLRQEHEDSTHR 527
Query 61 LKRQDKRRNLYSFGGDSGTAEAEEMYD--IEGGQRTTLVSRPATAYSQARAEAVENVQRV 118
LK +K+ L E EE++ +E +R ++ Q R A++ +
Sbjct 528 LKGLEKQYRL-------ARQEKEELHKQLVEASERLKSQTKELKDAHQQRKRALQEFSEL 580
Query 119 ---IGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQ 158
+ EL S+ Q+V+ + +EE ++ Q ID+ +R+ +
Sbjct 581 NERMSELRSLKQKVSRQLRDKEEEMEVAMQKIDSMRQDLRKSE 623
> cel:F57G4.1 hypothetical protein
Length=838
Score = 30.8 bits (68), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 38/147 (25%), Positives = 57/147 (38%), Gaps = 23/147 (15%)
Query 1 IRKTINTLNCKVDTLEKAAAAAGVSGQSK-QHYITVTEVLKTRLLD-------------V 46
+ K +TL+ K L+ A GQS QH I+V E L + LD +
Sbjct 112 LEKLKSTLSTKNPALKVNELAYSFIGQSNVQHAISVIETLNSEALDTIHFRTDVDEEINL 171
Query 47 TK-----EFKDILMLRTENLKRQDKRRNLYSFGGDSGTAEAEEMYDIEGGQRTTLVSRP- 100
TK + +I L+ E + N T + + D++ ++ L S+
Sbjct 172 TKIMELPHWPNITCLKIEGFIVSETLENFLHLTNAKVTKHSITIQDLQILKKNFLGSQSE 231
Query 101 ---ATAYSQARAEAVENVQRVIGELAS 124
Y E ENVQ V GEL+S
Sbjct 232 KKFVLVYKLDTGEIAENVQVVFGELSS 258
> dre:767671 ccdc146, MGC152979, si:ch211-276e22.2, zgc:152979;
coiled-coil domain containing 146
Length=860
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 41/85 (48%), Gaps = 12/85 (14%)
Query 98 SRPATAYSQARAEAVEN-VQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIRE 156
+P A ++R ++N + V E+ + Q V T+ + ++T HI++
Sbjct 167 DQPKQAELESRYATLKNSCEEVRKEVLKLRQEVRTLT-----------ESMETQQKHIKK 215
Query 157 GQNELLNFYNRISSNRSLIIKVFLL 181
Q EL + NR+ SN + + +V L+
Sbjct 216 EQLELEDLKNRVESNEAELAQVLLI 240
> hsa:55014 STX17, FLJ20651, MGC102796, MGC126613, MGC126615;
syntaxin 17; K08491 syntaxin 17
Length=302
Score = 30.0 bits (66), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 35/150 (23%), Positives = 64/150 (42%), Gaps = 19/150 (12%)
Query 12 VDTLEKAAAAAGVSGQSKQHYITVTEVLKTRLLDVTKEFKDILMLRTENLKRQDKRRNLY 71
+D +++ A+AA + + Q ++ E LK K+F D L L R
Sbjct 91 IDPVKEEASAA--TAEFLQLHLESVEELK-------KQFNDEETLLQPPLTRS------M 135
Query 72 SFGGDSGTAEAEEMYDIEGGQRTTLVSRPATAYSQARAEAVENVQRVIGELASIFQRVAT 131
+ GG T EAE T + + P Q AE+ E ++ + EL+ + +
Sbjct 136 TVGGAFHTTEAE----ASSQSLTQIYALPEIPQDQNAAESWETLEADLIELSQLVTDFSL 191
Query 132 MIAHQEEMIQRIGQDIDTSMYHIREGQNEL 161
++ Q+E I I ++++ ++ EG L
Sbjct 192 LVNSQQEKIDSIADHVNSAAVNVEEGTKNL 221
> ath:AT1G32270 ATSYP24; SNAP receptor/ protein binding; K08488
syntaxin 7
Length=416
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 32/61 (52%), Gaps = 1/61 (1%)
Query 106 QARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFY 165
+AR + ++ V+ I E+ +F+ +A M+ HQ I I + ID +G++ L+
Sbjct 269 EAREQGIQEVKHQISEVMEMFKDLAVMVDHQ-GTIDDIDEKIDNLRSAAAQGKSHLVKAS 327
Query 166 N 166
N
Sbjct 328 N 328
> cel:VF39H2L.1 hypothetical protein; K08491 syntaxin 17
Length=250
Score = 29.6 bits (65), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 12/56 (21%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 106 QARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNEL 161
+ RAEA +++ + +L IFQ + ++ Q +++ I + ++ + ++ G L
Sbjct 133 KERAEATVKIEKDMADLEKIFQELGRIVHEQHDVVDSIEEQVERATEDVKRGNENL 188
> mmu:67727 Stx17, 4833418L03Rik, 6330411F21Rik, 9030425C21Rik,
AW048351; syntaxin 17; K08491 syntaxin 17
Length=301
Score = 29.3 bits (64), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 15/68 (22%), Positives = 35/68 (51%), Gaps = 0/68 (0%)
Query 94 TTLVSRPATAYSQARAEAVENVQRVIGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYH 153
T + + P Q AE+ E ++ + EL+ + ++ +++ Q+E I I ++++ +
Sbjct 153 TQIYALPEIPQDQNAAESWETLEADLIELSHLVTDMSLLVSSQQEKIDSIADHVNSAAVN 212
Query 154 IREGQNEL 161
+ EG L
Sbjct 213 VEEGTKNL 220
> hsa:6809 STX3, FLJ30906, STX3A; syntaxin 3; K08486 syntaxin
1B/2/3
Length=277
Score = 29.3 bits (64), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 23/102 (22%), Positives = 49/102 (48%), Gaps = 10/102 (9%)
Query 67 RRNLYSFGGDSGTAEAEEMYDIEGGQRT--------TLVSRPATAYSQARAEAVENVQRV 118
+R L G + E EEM +E G + +S+ A + + R + + ++
Sbjct 150 QRQLEITGKKTTDEELEEM--LESGNPAIFTSGIIDSQISKQALSEIEGRHKDIVRLESS 207
Query 119 IGELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNE 160
I EL +F +A ++ +Q EM+ I ++ ++ H+ + ++E
Sbjct 208 IKELHDMFMDIAMLVENQGEMLDNIELNVMHTVDHVEKARDE 249
> dre:100150216 hypothetical LOC100150216
Length=1769
Score = 29.3 bits (64), Expect = 9.4, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 29/59 (49%), Gaps = 6/59 (10%)
Query 121 ELASIFQRVATMIAHQEEMIQRIGQDIDTSMYHIREGQNELLNFYNRISSNRSLIIKVF 179
E+AS FQ + HQE+ + + S+ ++ G NE+ R SS S +IK F
Sbjct 92 EMASTFQHSYVDVTHQEQAFR------EGSLPYMNNGSNEVTWQQGRSSSRVSSLIKAF 144
Lambda K H
0.321 0.134 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5608917492
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40