bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4034_orf1
Length=154
Score E
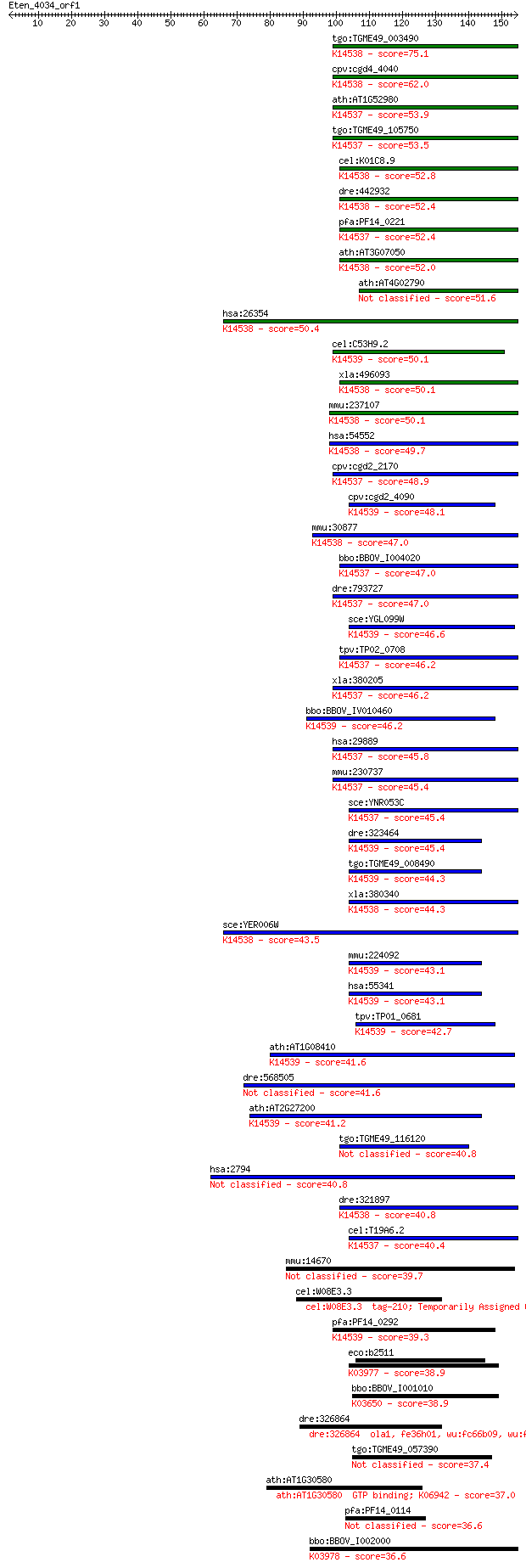
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_003490 GTPase domain containing protein ; K14538 nu... 75.1 8e-14
cpv:cgd4_4040 Yer006wp-like. Yjeq GTpase ; K14538 nuclear GTP-... 62.0 7e-10
ath:AT1G52980 GTP-binding family protein; K14537 nuclear GTP-b... 53.9 2e-07
tgo:TGME49_105750 nucleolar GTP-binding protein NOG2, putative... 53.5 2e-07
cel:K01C8.9 nst-1; mammalian NucleoSTemin (stem cell marker) r... 52.8 5e-07
dre:442932 gnl3l, MGC110536, SI:zK13A21.8, flj10613, flj10613l... 52.4 6e-07
pfa:PF14_0221 GTPase, putative; K14537 nuclear GTP-binding pro... 52.4 6e-07
ath:AT3G07050 GTP-binding family protein; K14538 nuclear GTP-b... 52.0 7e-07
ath:AT4G02790 GTP-binding family protein 51.6 1e-06
hsa:26354 GNL3, C77032, E2IG3, MGC800, NS; guanine nucleotide ... 50.4 2e-06
cel:C53H9.2 hypothetical protein; K14539 large subunit GTPase ... 50.1 3e-06
xla:496093 gnl3l; guanine nucleotide binding protein-like 3 (n... 50.1 3e-06
mmu:237107 Gnl3l, BC020354; guanine nucleotide binding protein... 50.1 3e-06
hsa:54552 GNL3L, FLJ10613; guanine nucleotide binding protein-... 49.7 4e-06
cpv:cgd2_2170 Ynr053p-like, Yjeq GTpase ; K14537 nuclear GTP-b... 48.9 7e-06
cpv:cgd2_4090 YawG/Kre35p-like, Yjeq GTpase ; K14539 large sub... 48.1 9e-06
mmu:30877 Gnl3, BC037996, C77032, MGC46970, Ns; guanine nucleo... 47.0 2e-05
bbo:BBOV_I004020 19.m02341; nucleolar GTP-binding protein 2; K... 47.0 2e-05
dre:793727 gnl2, MGC174441, MGC65800, MGC77319, wu:fc15c12, zg... 47.0 3e-05
sce:YGL099W LSG1, KRE35; Lsg1p; K14539 large subunit GTPase 1 ... 46.6 3e-05
tpv:TP02_0708 hypothetical protein; K14537 nuclear GTP-binding... 46.2 4e-05
xla:380205 gnl2, 1i973, MGC52791; guanine nucleotide binding p... 46.2 4e-05
bbo:BBOV_IV010460 23.m06487; GTPase subfamily protein; K14539 ... 46.2 4e-05
hsa:29889 GNL2, FLJ40906, HUMAUANTIG, NGP1, Ngp-1; guanine nuc... 45.8 6e-05
mmu:230737 Gnl2, BC003262, HUMAUANTIG, MGC7863, Ngp-1; guanine... 45.4 6e-05
sce:YNR053C NOG2, NUG2; Nog2p; K14537 nuclear GTP-binding protein 45.4 7e-05
dre:323464 fb99b06, lsg1, wu:fb99b06; zgc:76988 (EC:3.6.1.-); ... 45.4 8e-05
tgo:TGME49_008490 hypothetical protein ; K14539 large subunit ... 44.3 1e-04
xla:380340 gnl3, MGC53851, e2ig3, wu:fc55d07; guanine nucleoti... 44.3 2e-04
sce:YER006W NUG1; GTPase that associates with nuclear 60S pre-... 43.5 3e-04
mmu:224092 Lsg1, 5830465I20, AA409273, D16Bwg1547e; large subu... 43.1 3e-04
hsa:55341 LSG1, FLJ11301, FLJ27294; large subunit GTPase 1 hom... 43.1 3e-04
tpv:TP01_0681 hypothetical protein; K14539 large subunit GTPas... 42.7 4e-04
ath:AT1G08410 GTP-binding family protein; K14539 large subunit... 41.6 9e-04
dre:568505 gnl1, zgc:154008; guanine nucleotide binding protei... 41.6 0.001
ath:AT2G27200 GTP-binding family protein; K14539 large subunit... 41.2 0.001
tgo:TGME49_116120 GTP-binding protein, putative 40.8 0.002
hsa:2794 GNL1, DKFZp547E038, DKFZp547E128, FLJ37752, HSR1; gua... 40.8 0.002
dre:321897 gnl3, MGC123093, id:ibd2914, nstm, wu:fb38a04, wu:f... 40.8 0.002
cel:T19A6.2 ngp-1; Nuclear/nucleolar GTP-binding Protein famil... 40.4 0.002
mmu:14670 Gnl1, Gna-rs1, Gnal1; guanine nucleotide binding pro... 39.7 0.004
cel:W08E3.3 tag-210; Temporarily Assigned Gene name family mem... 39.3 0.005
pfa:PF14_0292 cytosolic preribosomal GTPase, putative; K14539 ... 39.3 0.005
eco:b2511 der, ECK2507, engA, JW5403, yfgK; GTPase; multicopy ... 38.9 0.006
bbo:BBOV_I001010 16.m00956; tRNA modification GTPase TrmE; K03... 38.9 0.007
dre:326864 ola1, fe36h01, wu:fc66b09, wu:fe36h01, wu:fk82a02, ... 37.7 0.014
tgo:TGME49_057390 GTP-binding conserved hypothetical domain-co... 37.4 0.019
ath:AT1G30580 GTP binding; K06942 37.0 0.026
pfa:PF14_0114 GTP binding protein, putative 36.6 0.035
bbo:BBOV_I002000 19.m02224; GTP-binding protein engB; K03978 G... 36.6 0.036
> tgo:TGME49_003490 GTPase domain containing protein ; K14538
nuclear GTP-binding protein
Length=673
Score = 75.1 bits (183), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 34/56 (60%), Positives = 46/56 (82%), Gaps = 0/56 (0%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
++KA + VGVVGYPNVGKSS+VNALT S A+VAA+PGST+ L+++++D QLI
Sbjct 300 TAKANLLVGVVGYPNVGKSSLVNALTHSCSAAAVAAMPGSTKTLQYVRIDKHIQLI 355
> cpv:cgd4_4040 Yer006wp-like. Yjeq GTpase ; K14538 nuclear GTP-binding
protein
Length=478
Score = 62.0 bits (149), Expect = 7e-10, Method: Composition-based stats.
Identities = 30/56 (53%), Positives = 41/56 (73%), Gaps = 1/56 (1%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
+SK +T+GV+GYPNVGKSS++N+L R C V A+ G TR L+ I LD+ T+LI
Sbjct 282 NSKKSITIGVMGYPNVGKSSLINSLKRGYCV-KVGAVAGVTRHLQRIDLDSTTKLI 336
> ath:AT1G52980 GTP-binding family protein; K14537 nuclear GTP-binding
protein
Length=576
Score = 53.9 bits (128), Expect = 2e-07, Method: Composition-based stats.
Identities = 29/57 (50%), Positives = 39/57 (68%), Gaps = 3/57 (5%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNAL-TRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
S K ++VG VGYPNVGKSSV+N L T++VC VA +PG T+ ++I L + LI
Sbjct 305 SDKQAISVGFVGYPNVGKSSVINTLRTKNVC--KVAPIPGETKVWQYITLTKRIFLI 359
> tgo:TGME49_105750 nucleolar GTP-binding protein NOG2, putative
; K14537 nuclear GTP-binding protein
Length=641
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 39/57 (68%), Gaps = 3/57 (5%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNAL-TRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
S + ++VG +GYPNVGKSS++NAL ++ VC A A +PG TR +++ L + LI
Sbjct 338 SDRKHVSVGFIGYPNVGKSSIINALRSKQVCRA--APIPGETRVWQYVALTKRLYLI 392
> cel:K01C8.9 nst-1; mammalian NucleoSTemin (stem cell marker)
related family member (nst-1); K14538 nuclear GTP-binding protein
Length=556
Score = 52.8 bits (125), Expect = 5e-07, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 38/54 (70%), Gaps = 1/54 (1%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
K + VGVVG+PNVGKSSV+N+L R +V LPG T++++ ++LD +LI
Sbjct 257 KTSIRVGVVGFPNVGKSSVINSLKRRK-ACNVGNLPGITKEIQEVELDKNIRLI 309
> dre:442932 gnl3l, MGC110536, SI:zK13A21.8, flj10613, flj10613l,
si:dkey-13a21.8, zgc:110536; guanine nucleotide binding
protein-like 3 (nucleolar)-like; K14538 nuclear GTP-binding
protein
Length=565
Score = 52.4 bits (124), Expect = 6e-07, Method: Composition-based stats.
Identities = 26/54 (48%), Positives = 38/54 (70%), Gaps = 1/54 (1%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
K +TVGVVG+PNVGKSS++N+L R+ +V A PG T+ L+ + LD +L+
Sbjct 241 KTAITVGVVGFPNVGKSSLINSLKRAR-ACNVGATPGVTKCLQEVHLDKHIKLL 293
> pfa:PF14_0221 GTPase, putative; K14537 nuclear GTP-binding protein
Length=487
Score = 52.4 bits (124), Expect = 6e-07, Method: Composition-based stats.
Identities = 24/54 (44%), Positives = 37/54 (68%), Gaps = 1/54 (1%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
K + +G++GYPNVGKS+++N+L + V S A +PG T+ +FIKL + LI
Sbjct 308 KKHIHIGLIGYPNVGKSAIINSLKKKVVCIS-ACIPGQTKYWQFIKLTNKIYLI 360
> ath:AT3G07050 GTP-binding family protein; K14538 nuclear GTP-binding
protein
Length=582
Score = 52.0 bits (123), Expect = 7e-07, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 37/54 (68%), Gaps = 1/54 (1%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
K +TVG++G PNVGKSS++N+L R+ +V A PG TR L+ + LD +L+
Sbjct 251 KKSITVGIIGLPNVGKSSLINSLKRA-HVVNVGATPGLTRSLQEVHLDKNVKLL 303
> ath:AT4G02790 GTP-binding family protein
Length=372
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 34/49 (69%), Gaps = 3/49 (6%)
Query 107 GVVGYPNVGKSSVVN-ALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
G++GYPNVGKSS++N L R +C A A PG TR++K++KL L+
Sbjct 217 GIIGYPNVGKSSLINRLLKRKICAA--APRPGVTREMKWVKLGKDLDLL 263
> hsa:26354 GNL3, C77032, E2IG3, MGC800, NS; guanine nucleotide
binding protein-like 3 (nucleolar); K14538 nuclear GTP-binding
protein
Length=549
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 32/101 (31%), Positives = 51/101 (50%), Gaps = 14/101 (13%)
Query 66 AATSTAEGDKITTESRASATSSSSRPA-----------LGGGSGSSKAFMTVGVVGYPNV 114
A+T + KIT +A ++ R LGG + + VGV+G+PNV
Sbjct 206 ASTKPKDKGKITKRVKAKKNAAPFRSEVCFGKEGLWKLLGGFQETCSKAIRVGVIGFPNV 265
Query 115 GKSSVVNALTR-SVCTASVAALPGSTRQLKFIKLDAQTQLI 154
GKSS++N+L + +C V+ G TR ++ + LD Q +I
Sbjct 266 GKSSIINSLKQEQMCNVGVSM--GLTRSMQVVPLDKQITII 304
> cel:C53H9.2 hypothetical protein; K14539 large subunit GTPase
1 [EC:3.6.1.-]
Length=554
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/52 (51%), Positives = 34/52 (65%), Gaps = 2/52 (3%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQ 150
S+K M VG+VGYPNVGKSS +N L SV+A PG TR + I +D+Q
Sbjct 295 SAKPVM-VGMVGYPNVGKSSTINKLAGGK-KVSVSATPGKTRHFQTIHIDSQ 344
> xla:496093 gnl3l; guanine nucleotide binding protein-like 3
(nucleolar)-like; K14538 nuclear GTP-binding protein
Length=561
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 25/54 (46%), Positives = 38/54 (70%), Gaps = 1/54 (1%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
K ++VGVVG+PNVGKSS++N+L R+ +V A PG T+ L+ + LD +L+
Sbjct 241 KTSISVGVVGFPNVGKSSLINSLKRAR-ACNVGATPGVTKCLQEVHLDKHIKLL 293
> mmu:237107 Gnl3l, BC020354; guanine nucleotide binding protein-like
3 (nucleolar)-like; K14538 nuclear GTP-binding protein
Length=577
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 98 GSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
G + + VGVVG PNVGKSS++N+L RS SV A+PG T+ ++ + LD +L+
Sbjct 241 GEVRGHIRVGVVGLPNVGKSSLINSLKRSR-ACSVGAVPGVTKFMQEVYLDKFIRLL 296
> hsa:54552 GNL3L, FLJ10613; guanine nucleotide binding protein-like
3 (nucleolar)-like; K14538 nuclear GTP-binding protein
Length=582
Score = 49.7 bits (117), Expect = 4e-06, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 98 GSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
G + + VGVVG PNVGKSS++N+L RS SV A+PG T+ ++ + LD +L+
Sbjct 247 GEVRTHIRVGVVGLPNVGKSSLINSLKRSR-ACSVGAVPGITKFMQEVYLDKFIRLL 302
> cpv:cgd2_2170 Ynr053p-like, Yjeq GTpase ; K14537 nuclear GTP-binding
protein
Length=562
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 27/57 (47%), Positives = 37/57 (64%), Gaps = 3/57 (5%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNALTRS-VCTASVAALPGSTRQLKFIKLDAQTQLI 154
S K ++VG +GYPNVGKSS++N L S VC SVA + G T+ ++I L + LI
Sbjct 333 SDKKHVSVGFIGYPNVGKSSIINTLRGSKVC--SVAPIAGETKIWQYIHLTHRIYLI 387
> cpv:cgd2_4090 YawG/Kre35p-like, Yjeq GTpase ; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=666
Score = 48.1 bits (113), Expect = 9e-06, Method: Composition-based stats.
Identities = 22/44 (50%), Positives = 34/44 (77%), Gaps = 1/44 (2%)
Query 104 MTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKL 147
+T+G+VG+PNVGKSS+VNAL S +S++ PG T+ L+ ++L
Sbjct 417 LTIGMVGFPNVGKSSIVNALFGSQ-KSSISRTPGKTKHLQTLRL 459
> mmu:30877 Gnl3, BC037996, C77032, MGC46970, Ns; guanine nucleotide
binding protein-like 3 (nucleolar); K14538 nuclear GTP-binding
protein
Length=538
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 38/63 (60%), Gaps = 3/63 (4%)
Query 93 LGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTRS-VCTASVAALPGSTRQLKFIKLDAQT 151
LG S + VGV+G+PNVGKSSV+N+L + +C ++ G TR ++ + LD Q
Sbjct 239 LGDFQQSCGKDIQVGVIGFPNVGKSSVINSLKQEWICNVGISM--GLTRSMQIVPLDKQI 296
Query 152 QLI 154
+I
Sbjct 297 TII 299
> bbo:BBOV_I004020 19.m02341; nucleolar GTP-binding protein 2;
K14537 nuclear GTP-binding protein
Length=671
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/55 (43%), Positives = 33/55 (60%), Gaps = 3/55 (5%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALT-RSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
+ +VG +GYPNVGKSSV+N L C A A +PG TR +++ L + LI
Sbjct 306 RKHFSVGFIGYPNVGKSSVINTLKGEKNCKA--APIPGETRVWQYVSLTKRIHLI 358
> dre:793727 gnl2, MGC174441, MGC65800, MGC77319, wu:fc15c12,
zgc:65800, zgc:77319; guanine nucleotide binding protein-like
2 (nucleolar); K14537 nuclear GTP-binding protein
Length=727
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/56 (46%), Positives = 36/56 (64%), Gaps = 1/56 (1%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
S K ++VG +GYPNVGKSS++N L RS +VA L G T+ ++I L + LI
Sbjct 306 SDKKQISVGFIGYPNVGKSSIINTL-RSKKVCNVAPLAGETKVWQYITLMRRIFLI 360
> sce:YGL099W LSG1, KRE35; Lsg1p; K14539 large subunit GTPase
1 [EC:3.6.1.-]
Length=640
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 22/50 (44%), Positives = 32/50 (64%), Gaps = 1/50 (2%)
Query 104 MTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQL 153
+ +G+VGYPNVGKSS +N+L + SV++ PG T+ + IKL L
Sbjct 337 INIGLVGYPNVGKSSTINSLVGAK-KVSVSSTPGKTKHFQTIKLSDSVML 385
> tpv:TP02_0708 hypothetical protein; K14537 nuclear GTP-binding
protein
Length=556
Score = 46.2 bits (108), Expect = 4e-05, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 34/56 (60%), Gaps = 5/56 (8%)
Query 101 KAFMTVGVVGYPNVGKSSVVNAL--TRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
+ +VG +GYPNVGKSSV+N L RS TA + PG TR +++ L + LI
Sbjct 305 RKHFSVGFIGYPNVGKSSVINTLKGNRSCKTAPI---PGETRVWQYVCLTKRIHLI 357
> xla:380205 gnl2, 1i973, MGC52791; guanine nucleotide binding
protein-like 2 (nucleolar); K14537 nuclear GTP-binding protein
Length=707
Score = 46.2 bits (108), Expect = 4e-05, Method: Composition-based stats.
Identities = 25/57 (43%), Positives = 38/57 (66%), Gaps = 3/57 (5%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNAL-TRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
+ K ++VG +GYPNVGKSSV+N L ++ VC +VA + G T+ ++I L + LI
Sbjct 306 TDKKQISVGFIGYPNVGKSSVINTLRSKKVC--NVAPIAGETKVWQYITLMRRIFLI 360
> bbo:BBOV_IV010460 23.m06487; GTPase subfamily protein; K14539
large subunit GTPase 1 [EC:3.6.1.-]
Length=826
Score = 46.2 bits (108), Expect = 4e-05, Method: Composition-based stats.
Identities = 25/57 (43%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 91 PALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKL 147
P L S TVG VGYPNVGKSS++N L V +V+ PG T+ L+ + L
Sbjct 549 PELRQEDTSDMPVYTVGCVGYPNVGKSSLINCLME-VTKTNVSCQPGKTKHLQTLAL 604
> hsa:29889 GNL2, FLJ40906, HUMAUANTIG, NGP1, Ngp-1; guanine nucleotide
binding protein-like 2 (nucleolar); K14537 nuclear
GTP-binding protein
Length=731
Score = 45.8 bits (107), Expect = 6e-05, Method: Composition-based stats.
Identities = 25/57 (43%), Positives = 38/57 (66%), Gaps = 3/57 (5%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNAL-TRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
+ K ++VG +GYPNVGKSSV+N L ++ VC +VA + G T+ ++I L + LI
Sbjct 306 TDKKQISVGFIGYPNVGKSSVINTLRSKKVC--NVAPIAGETKVWQYITLMRRIFLI 360
> mmu:230737 Gnl2, BC003262, HUMAUANTIG, MGC7863, Ngp-1; guanine
nucleotide binding protein-like 2 (nucleolar); K14537 nuclear
GTP-binding protein
Length=728
Score = 45.4 bits (106), Expect = 6e-05, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 36/56 (64%), Gaps = 1/56 (1%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
+ K ++VG +GYPNVGKSSV+N L RS +VA + G T+ ++I L + LI
Sbjct 306 TDKKQISVGFIGYPNVGKSSVINTL-RSKKVCNVAPIAGETKVWQYITLMRRIFLI 360
> sce:YNR053C NOG2, NUG2; Nog2p; K14537 nuclear GTP-binding protein
Length=486
Score = 45.4 bits (106), Expect = 7e-05, Method: Composition-based stats.
Identities = 23/52 (44%), Positives = 34/52 (65%), Gaps = 3/52 (5%)
Query 104 MTVGVVGYPNVGKSSVVNAL-TRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
++VG +GYPN GKSS++N L + VC VA +PG T+ ++I L + LI
Sbjct 316 ISVGFIGYPNTGKSSIINTLRKKKVC--QVAPIPGETKVWQYITLMKRIFLI 365
> dre:323464 fb99b06, lsg1, wu:fb99b06; zgc:76988 (EC:3.6.1.-);
K14539 large subunit GTPase 1 [EC:3.6.1.-]
Length=640
Score = 45.4 bits (106), Expect = 8e-05, Method: Composition-based stats.
Identities = 21/40 (52%), Positives = 29/40 (72%), Gaps = 1/40 (2%)
Query 104 MTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLK 143
+TVG+VGYPNVGKSS +N + R+ SV+A PG T+ +
Sbjct 369 ITVGLVGYPNVGKSSTINTIFRNK-KVSVSATPGHTKHFQ 407
> tgo:TGME49_008490 hypothetical protein ; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=1064
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/40 (57%), Positives = 28/40 (70%), Gaps = 1/40 (2%)
Query 104 MTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLK 143
VG+VG+PNVGKSSV+NAL S SV+ PG TR L+
Sbjct 667 FIVGLVGFPNVGKSSVINALLGSK-KVSVSRTPGKTRHLQ 705
> xla:380340 gnl3, MGC53851, e2ig3, wu:fc55d07; guanine nucleotide
binding protein-like 3 (nucleolar); K14538 nuclear GTP-binding
protein
Length=542
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 24/52 (46%), Positives = 38/52 (73%), Gaps = 3/52 (5%)
Query 104 MTVGVVGYPNVGKSSVVNALTRS-VCTASVAALPGSTRQLKFIKLDAQTQLI 154
+ VGV+G+ NVGKSSV+N+L +S VC +V G+T+ L+ ++LD Q +L+
Sbjct 248 IKVGVIGFANVGKSSVINSLKQSHVC--NVGPSKGTTKFLQEVRLDPQIRLL 297
> sce:YER006W NUG1; GTPase that associates with nuclear 60S pre-ribosomes,
required for export of 60S ribosomal subunits from
the nucleus; K14538 nuclear GTP-binding protein
Length=520
Score = 43.5 bits (101), Expect = 3e-04, Method: Composition-based stats.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 6/95 (6%)
Query 66 AATSTAEGDKITTESRASATSSSSRPALGGGSGSS--KAFMTVGVVGYPNVGKSSVVNAL 123
A++ G + + T+S+ +L S +S K + VGV+GYPNVGKSSV+NAL
Sbjct 241 ASSGAVNGTSFNRKLSQTTTASALLESLKTYSNNSNLKRSIVVGVIGYPNVGKSSVINAL 300
Query 124 TRSVCTAS----VAALPGSTRQLKFIKLDAQTQLI 154
S V G T L+ IK+D + +++
Sbjct 301 LARRGGQSKACPVGNEAGVTTSLREIKIDNKLKIL 335
> mmu:224092 Lsg1, 5830465I20, AA409273, D16Bwg1547e; large subunit
GTPase 1 homolog (S. cerevisiae); K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=644
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 20/40 (50%), Positives = 28/40 (70%), Gaps = 1/40 (2%)
Query 104 MTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLK 143
+TVG+VGYPNVGKSS +N + + SV+A PG T+ +
Sbjct 373 LTVGLVGYPNVGKSSTINTIMGNK-KVSVSATPGHTKHFQ 411
> hsa:55341 LSG1, FLJ11301, FLJ27294; large subunit GTPase 1 homolog
(S. cerevisiae); K14539 large subunit GTPase 1 [EC:3.6.1.-]
Length=658
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 20/40 (50%), Positives = 28/40 (70%), Gaps = 1/40 (2%)
Query 104 MTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLK 143
+TVG+VGYPNVGKSS +N + + SV+A PG T+ +
Sbjct 387 LTVGLVGYPNVGKSSTINTIMGNK-KVSVSATPGHTKHFQ 425
> tpv:TP01_0681 hypothetical protein; K14539 large subunit GTPase
1 [EC:3.6.1.-]
Length=529
Score = 42.7 bits (99), Expect = 4e-04, Method: Composition-based stats.
Identities = 21/42 (50%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 106 VGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKL 147
VG VGYPNVGKSS++N L S T V PG T+ ++ + L
Sbjct 447 VGFVGYPNVGKSSLINCLMESTRTC-VGTQPGKTKHIQTLPL 487
> ath:AT1G08410 GTP-binding family protein; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=589
Score = 41.6 bits (96), Expect = 9e-04, Method: Composition-based stats.
Identities = 29/74 (39%), Positives = 39/74 (52%), Gaps = 6/74 (8%)
Query 80 SRASATSSSSRPALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGST 139
SRA++ SS S G +A VG VGYPNVGKSS +NAL T V + PG T
Sbjct 290 SRAASVSSQS---WTGEYQRDQA--VVGFVGYPNVGKSSTINALVGQKRTG-VTSTPGKT 343
Query 140 RQLKFIKLDAQTQL 153
+ + + + + L
Sbjct 344 KHFQTLIISDELML 357
> dre:568505 gnl1, zgc:154008; guanine nucleotide binding protein-like
1
Length=602
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 45/87 (51%), Gaps = 7/87 (8%)
Query 72 EGDKITTESRASATSSSSRPALGGGSGSSKAF----MTVGVVGYPNVGKSSVVNALT-RS 126
EGD+ S + S A+ S + + + +T+G +G+PNVGKSSV+N+L R
Sbjct 313 EGDRADDGSESVLMEHHSDIAMEMNSPTQELYKDGVLTLGCIGFPNVGKSSVLNSLVGRK 372
Query 127 VCTASVAALPGSTRQLKFIKLDAQTQL 153
V SV+ PG T+ + L +L
Sbjct 373 V--VSVSRTPGHTKYFQTYYLTPTVKL 397
> ath:AT2G27200 GTP-binding family protein; K14539 large subunit
GTPase 1 [EC:3.6.1.-]
Length=537
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 34/70 (48%), Gaps = 1/70 (1%)
Query 74 DKITTESRASATSSSSRPALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVA 133
D++ E+ SR + S + VG VGYPNVGKSS +NAL T V
Sbjct 275 DRLKLEALEIVKMRKSRGVSATSTESHCEQVVVGFVGYPNVGKSSTINALVGQKRTG-VT 333
Query 134 ALPGSTRQLK 143
+ PG T+ +
Sbjct 334 STPGKTKHFQ 343
> tgo:TGME49_116120 GTP-binding protein, putative
Length=733
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/39 (48%), Positives = 27/39 (69%), Gaps = 2/39 (5%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGST 139
K+ VG+VG+PN GKSS++ AL+R C VA+ P +T
Sbjct 227 KSIADVGLVGFPNAGKSSILAALSR--CAPRVASFPFTT 263
> hsa:2794 GNL1, DKFZp547E038, DKFZp547E128, FLJ37752, HSR1; guanine
nucleotide binding protein-like 1
Length=607
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 42/93 (45%), Gaps = 6/93 (6%)
Query 62 HGGAAATSTAEGDKITTESRASATSSSSRPALGGGSGSSKAFMTVGVVGYPNVGKSSVVN 121
+G +G + E + T S+ P +T+G VG+PNVGKSS++N
Sbjct 322 NGSGEEEEEEDGPAVLVEQQ---TDSAMEPTGPTQERYKDGVVTIGCVGFPNVGKSSLIN 378
Query 122 ALT-RSVCTASVAALPGSTRQLKFIKLDAQTQL 153
L R V SV+ PG TR + L +L
Sbjct 379 GLVGRKV--VSVSRTPGHTRYFQTYFLTPSVKL 409
> dre:321897 gnl3, MGC123093, id:ibd2914, nstm, wu:fb38a04, wu:fc55d07,
zgc:123093; guanine nucleotide binding protein-like
3 (nucleolar); K14538 nuclear GTP-binding protein
Length=561
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 34/55 (61%), Gaps = 3/55 (5%)
Query 101 KAFMTVGVVGYPNVGKSSVVNALTR-SVCTASVAALPGSTRQLKFIKLDAQTQLI 154
+ + VGVVG+PNVGKSS++N+L C A V G TR ++ + + + ++I
Sbjct 259 ETMLKVGVVGFPNVGKSSIINSLKEMRACNAGVQR--GLTRCMQEVHITKKVKMI 311
> cel:T19A6.2 ngp-1; Nuclear/nucleolar GTP-binding Protein family
member (ngp-1); K14537 nuclear GTP-binding protein
Length=651
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 22/52 (42%), Positives = 33/52 (63%), Gaps = 3/52 (5%)
Query 104 MTVGVVGYPNVGKSSVVNAL-TRSVCTASVAALPGSTRQLKFIKLDAQTQLI 154
++VG +GYPNVGKSS+VN L + VC A + G T+ +++ L + LI
Sbjct 326 ISVGFIGYPNVGKSSLVNTLRKKKVC--KTAPIAGETKVWQYVMLMRRIYLI 375
> mmu:14670 Gnl1, Gna-rs1, Gnal1; guanine nucleotide binding protein-like
1
Length=607
Score = 39.7 bits (91), Expect = 0.004, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query 85 TSSSSRPALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALT-RSVCTASVAALPGSTRQLK 143
T S+ P +T+G +G+PNVGKSS++N L R V SV+ PG TR +
Sbjct 342 TDSAMEPTGPSRERYKDGVVTIGCIGFPNVGKSSLINGLVGRKV--VSVSRTPGHTRYFQ 399
Query 144 FIKLDAQTQL 153
L +L
Sbjct 400 TYFLTPSVKL 409
> cel:W08E3.3 tag-210; Temporarily Assigned Gene name family member
(tag-210); K06942
Length=395
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 88 SSRPALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTAS 131
+PAL G G++ + VG++G PNVGKS+ N LT+S A
Sbjct 9 EEKPALIGRLGTN---LKVGILGLPNVGKSTFFNVLTKSEAQAE 49
> pfa:PF14_0292 cytosolic preribosomal GTPase, putative; K14539
large subunit GTPase 1 [EC:3.6.1.-]
Length=833
Score = 39.3 bits (90), Expect = 0.005, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 99 SSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKL 147
SS +G +G+PNVGKSS++N L SV+ PG T+ + I L
Sbjct 594 SSIPKFMIGFIGFPNVGKSSIINCLIGKK-KVSVSRQPGKTKHFQTITL 641
> eco:b2511 der, ECK2507, engA, JW5403, yfgK; GTPase; multicopy
suppressor of FtsJ; K03977 GTP-binding protein
Length=490
Score = 38.9 bits (89), Expect = 0.006, Method: Composition-based stats.
Identities = 21/39 (53%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 106 VGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKF 144
V +VG PNVGKS++ N LTR+ A VA PG TR K+
Sbjct 5 VALVGRPNVGKSTLFNRLTRTR-DALVADFPGLTRDRKY 42
Score = 30.0 bits (66), Expect = 3.2, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 7/48 (14%)
Query 104 MTVGVVGYPNVGKSSVVNAL---TRSVCTASVAALPGSTRQLKFIKLD 148
+ + +VG PNVGKS++ N + R V V +PG+TR +I ++
Sbjct 203 IKLAIVGRPNVGKSTLTNRILGEERVV----VYDMPGTTRDSIYIPME 246
> bbo:BBOV_I001010 16.m00956; tRNA modification GTPase TrmE; K03650
tRNA modification GTPase
Length=529
Score = 38.9 bits (89), Expect = 0.007, Method: Composition-based stats.
Identities = 23/45 (51%), Positives = 29/45 (64%), Gaps = 3/45 (6%)
Query 105 TVGVVGYPNVGKSSVVNALT-RSVCTASVAALPGSTRQLKFIKLD 148
TV +VG PN GKSS++N + R+V A VA LPG+TR K D
Sbjct 275 TVAIVGPPNAGKSSLINTICDRNV--AIVADLPGTTRDPVHAKYD 317
> dre:326864 ola1, fe36h01, wu:fc66b09, wu:fe36h01, wu:fk82a02,
zgc:55768, zgc:85691; Obg-like ATPase 1 (EC:3.6.3.-); K06942
Length=396
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 26/43 (60%), Gaps = 3/43 (6%)
Query 89 SRPALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTAS 131
+P L G G+S + +G+VG PNVGKS+ N LT+S A
Sbjct 11 KQPPLIGRFGTS---LKIGIVGLPNVGKSTFFNVLTKSQAAAE 50
> tgo:TGME49_057390 GTP-binding conserved hypothetical domain-containing
protein
Length=1466
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 105 TVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIK 146
+ G NVGKSS++N L T+ V + PG+T+QL F K
Sbjct 457 EIAFAGRSNVGKSSLLNELAGRRLTSLVTSRPGTTQQLHFFK 498
> ath:AT1G30580 GTP binding; K06942
Length=394
Score = 37.0 bits (84), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 4/47 (8%)
Query 79 ESRASATSSSSRPALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTR 125
+++A RP LG S K +G+VG PNVGKS++ N LT+
Sbjct 4 KAKAKDAGPVERPILGRFSSHLK----IGIVGLPNVGKSTLFNTLTK 46
> pfa:PF14_0114 GTP binding protein, putative
Length=627
Score = 36.6 bits (83), Expect = 0.035, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 21/24 (87%), Gaps = 0/24 (0%)
Query 103 FMTVGVVGYPNVGKSSVVNALTRS 126
F +G++GYPNVGKS+++N +T++
Sbjct 191 FTDLGIIGYPNVGKSTLLNKITKA 214
> bbo:BBOV_I002000 19.m02224; GTP-binding protein engB; K03978
GTP-binding protein
Length=290
Score = 36.6 bits (83), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 0/63 (0%)
Query 92 ALGGGSGSSKAFMTVGVVGYPNVGKSSVVNALTRSVCTASVAALPGSTRQLKFIKLDAQT 151
A+G A V G N GKS ++NA+ V LPG+TR+L F K+ +
Sbjct 45 AIGHNDLPPAALPEVAFYGRTNAGKSCLINAICGRYGVCPVHELPGTTRKLHFYKVGTPS 104
Query 152 QLI 154
+I
Sbjct 105 TVI 107
Lambda K H
0.315 0.127 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40