bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3954_orf3
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
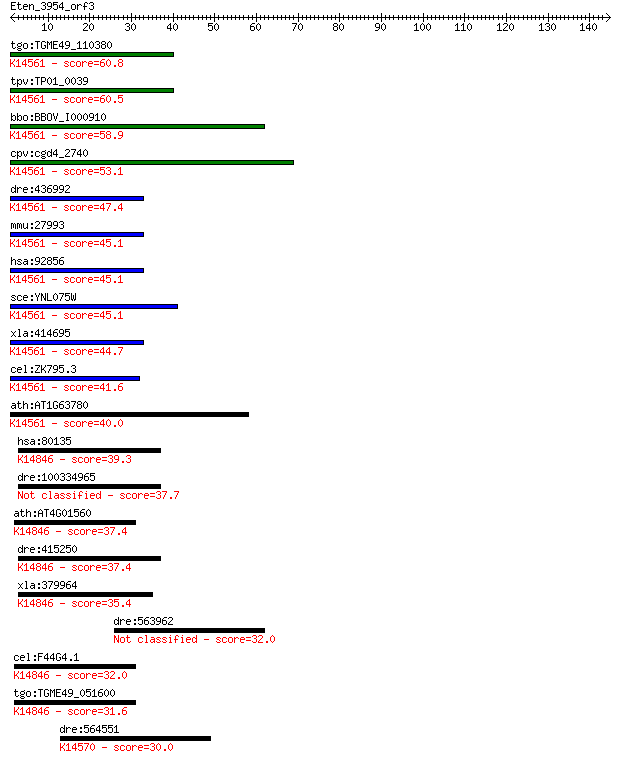
tgo:TGME49_110380 U3 small nucleolar ribonucleoprotein protein... 60.8 2e-09
tpv:TP01_0039 U3 small nuclear ribonucleoprotein; K14561 U3 sm... 60.5 2e-09
bbo:BBOV_I000910 16.m00780; Brix domain containing protein; K1... 58.9 5e-09
cpv:cgd4_2740 IMP4 U3 small nucleolar ribonucleoprotein ; K145... 53.1 3e-07
dre:436992 imp4, zgc:86740; IMP4, U3 small nucleolar ribonucle... 47.4 2e-05
mmu:27993 Imp4, AA409888, AV031295, D1Wsu40e; IMP4, U3 small n... 45.1 8e-05
hsa:92856 IMP4, BXDC4, MGC19606; IMP4, U3 small nucleolar ribo... 45.1 8e-05
sce:YNL075W IMP4; Component of the SSU processome, which is re... 45.1 8e-05
xla:414695 imp4, MGC83136; IMP4, U3 small nucleolar ribonucleo... 44.7 9e-05
cel:ZK795.3 hypothetical protein; K14561 U3 small nucleolar ri... 41.6 8e-04
ath:AT1G63780 IMP4; IMP4; K14561 U3 small nucleolar ribonucleo... 40.0 0.003
hsa:80135 RPF1, BXDC5, DKFZp761G0415, DKFZp761M0215, FLJ12475,... 39.3 0.004
dre:100334965 ribosome production factor 1-like 37.7 0.013
ath:AT4G01560 MEE49; MEE49 (maternal effect embryo arrest 49);... 37.4 0.015
dre:415250 rpf1, bxdc5, zgc:86604; ribosome production factor ... 37.4 0.016
xla:379964 bxdc5, MGC53934, rpf1; RNA processing factor 1; K14... 35.4 0.069
dre:563962 slc26a3, si:dkey-31f5.2; solute carrier family 26, ... 32.0 0.60
cel:F44G4.1 hypothetical protein; K14846 ribosome production f... 32.0 0.74
tgo:TGME49_051600 RNA processing factor, putative ; K14846 rib... 31.6 0.92
dre:564551 rexo1, fb77b11, si:dkey-45m5.5, wu:fb77b11; REX1, R... 30.0 2.3
> tgo:TGME49_110380 U3 small nucleolar ribonucleoprotein protein,
putative (EC:3.1.2.15); K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=314
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 26/39 (66%), Positives = 31/39 (79%), Gaps = 0/39 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPNMSQ 39
GQP ++V HLPHGP A+F LSGV LRHDLPE+ NMS+
Sbjct 145 GQPDAMVVCHLPHGPAAYFNLSGVALRHDLPEKPANMSE 183
> tpv:TP01_0039 U3 small nuclear ribonucleoprotein; K14561 U3
small nucleolar ribonucleoprotein protein IMP4
Length=302
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 25/39 (64%), Positives = 29/39 (74%), Gaps = 0/39 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPNMSQ 39
G P +I+ HLPHGPTA+F LS V LRHDL E+ P MSQ
Sbjct 154 GNPNSMIITHLPHGPTAYFQLSDVVLRHDLTEKLPTMSQ 192
> bbo:BBOV_I000910 16.m00780; Brix domain containing protein;
K14561 U3 small nucleolar ribonucleoprotein protein IMP4
Length=293
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 38/65 (58%), Gaps = 4/65 (6%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPNMSQ----VLFCCCCSAAAAAAAAL 56
G+P +I+ HLPHGPTA+F LS V LRHDLPE+ P MS+ ++F S +
Sbjct 144 GEPNAMIICHLPHGPTAYFQLSQVVLRHDLPEKPPTMSEAYPHLIFHNFSSQLGERVKNI 203
Query 57 LLLPF 61
+ F
Sbjct 204 IRYMF 208
> cpv:cgd4_2740 IMP4 U3 small nucleolar ribonucleoprotein ; K14561
U3 small nucleolar ribonucleoprotein protein IMP4
Length=288
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 40/72 (55%), Gaps = 4/72 (5%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPNMSQ----VLFCCCCSAAAAAAAAL 56
G+P LI+ H P+GPTA+F +S V LRHDLP + MSQ ++F S + +
Sbjct 143 GEPDGLIISHFPYGPTAYFAISDVVLRHDLPIKPSTMSQSIPHLIFQNFNSTLGLRVSNI 202
Query 57 LLLPFWRLLRAA 68
L F +L A+
Sbjct 203 LKHLFPKLKNAS 214
> dre:436992 imp4, zgc:86740; IMP4, U3 small nucleolar ribonucleoprotein,
homolog (yeast); K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=291
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 19/32 (59%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPE 32
GQP L+V HLP GPTA+FT+ V +RHD+P+
Sbjct 144 GQPDGLVVCHLPFGPTAYFTMYNVVMRHDVPD 175
> mmu:27993 Imp4, AA409888, AV031295, D1Wsu40e; IMP4, U3 small
nucleolar ribonucleoprotein, homolog (yeast); K14561 U3 small
nucleolar ribonucleoprotein protein IMP4
Length=291
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPE 32
G P LIV HLP GPTA+FTL V +RHD+P+
Sbjct 144 GTPVGLIVSHLPFGPTAYFTLCNVVMRHDIPD 175
> hsa:92856 IMP4, BXDC4, MGC19606; IMP4, U3 small nucleolar ribonucleoprotein,
homolog (yeast); K14561 U3 small nucleolar
ribonucleoprotein protein IMP4
Length=291
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPE 32
G P LIV HLP GPTA+FTL V +RHD+P+
Sbjct 144 GTPVGLIVSHLPFGPTAYFTLCNVVMRHDIPD 175
> sce:YNL075W IMP4; Component of the SSU processome, which is
required for pre-18S rRNA processing; interacts with Mpp10p;
member of a superfamily of proteins that contain a sigma(70)-like
motif and associate with RNAs; K14561 U3 small nucleolar
ribonucleoprotein protein IMP4
Length=290
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/40 (50%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPNMSQV 40
G P L + H PHGPTA F+L V +RHD+ A N S+V
Sbjct 147 GVPTSLTISHFPHGPTAQFSLHNVVMRHDIIN-AGNQSEV 185
> xla:414695 imp4, MGC83136; IMP4, U3 small nucleolar ribonucleoprotein,
homolog; K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=291
Score = 44.7 bits (104), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 20/32 (62%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLPE 32
G P LIV HLP GPTA+FTL V +RHD+P+
Sbjct 144 GMPDGLIVCHLPFGPTAYFTLCNVVMRHDIPD 175
> cel:ZK795.3 hypothetical protein; K14561 U3 small nucleolar
ribonucleoprotein protein IMP4
Length=292
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHDLP 31
G P ++V HLP GPTA F+++ V +RHD+P
Sbjct 139 GNPDGMLVCHLPFGPTAFFSMANVVMRHDIP 169
> ath:AT1G63780 IMP4; IMP4; K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=294
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 31/66 (46%), Gaps = 12/66 (18%)
Query 1 GQPPRLIVFHLPHGPTAHFTLSGVCLRHD---------LPERAPNMSQVLFCCCCSAAAA 51
G P LI+ HLP GPTA+F L V RHD +PE+ P++ +F +
Sbjct 144 GVPDGLIISHLPFGPTAYFGLLNVVTRHDISDKKSIGKMPEQYPHL---IFNNFTTQMGQ 200
Query 52 AAAALL 57
+L
Sbjct 201 RVGNIL 206
> hsa:80135 RPF1, BXDC5, DKFZp761G0415, DKFZp761M0215, FLJ12475,
RP11-118B23.1; ribosome production factor 1 homolog (S. cerevisiae);
K14846 ribosome production factor 1
Length=349
Score = 39.3 bits (90), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 3 PPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPN 36
P LI+ HLP+GPTAHF +S V LR ++ R +
Sbjct 205 PNGLILSHLPNGPTAHFKMSSVRLRKEIKRRGKD 238
> dre:100334965 ribosome production factor 1-like
Length=352
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 3 PPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPN 36
P +++ HLP GPTAHF +S V LR ++ R N
Sbjct 208 PNGMVLCHLPDGPTAHFKVSSVRLRKEMKRRGKN 241
> ath:AT4G01560 MEE49; MEE49 (maternal effect embryo arrest 49);
K14846 ribosome production factor 1
Length=343
Score = 37.4 bits (85), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 2 QPPRLIVFHLPHGPTAHFTLSGVCLRHDL 30
+P L++ LP+GPTAHF LS + LR D+
Sbjct 189 EPDALLIIGLPNGPTAHFKLSNLVLRKDI 217
> dre:415250 rpf1, bxdc5, zgc:86604; ribosome production factor
1 homolog (S. cerevisiae); K14846 ribosome production factor
1
Length=330
Score = 37.4 bits (85), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 3 PPRLIVFHLPHGPTAHFTLSGVCLRHDLPERAPN 36
P +++ HLP GPTAHF +S V LR ++ R N
Sbjct 186 PNGMVLCHLPDGPTAHFKVSSVRLRKEMKRRGKN 219
> xla:379964 bxdc5, MGC53934, rpf1; RNA processing factor 1; K14846
ribosome production factor 1
Length=343
Score = 35.4 bits (80), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 3 PPRLIVFHLPHGPTAHFTLSGVCLRHDLPERA 34
P L++ HLP GPTAHF +S V LR ++ +
Sbjct 199 PNGLVLCHLPDGPTAHFKISNVRLRKEMKRKG 230
> dre:563962 slc26a3, si:dkey-31f5.2; solute carrier family 26,
member 3
Length=745
Score = 32.0 bits (71), Expect = 0.60, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Query 26 LRHDLPE--RAPNMSQVLFCCCCSAAAAAAAALLLLPF 61
LRH L + + S++ CC C A A AAL L PF
Sbjct 34 LRHHLKDYFSGVSFSKLCLCCSCDAKRAKNAALSLFPF 71
> cel:F44G4.1 hypothetical protein; K14846 ribosome production
factor 1
Length=384
Score = 32.0 bits (71), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 1/30 (3%)
Query 2 QPPRLIVF-HLPHGPTAHFTLSGVCLRHDL 30
+ P I+F HLP GPTA+F ++ + DL
Sbjct 238 KKPNGIIFCHLPEGPTAYFKINSLTFTQDL 267
> tgo:TGME49_051600 RNA processing factor, putative ; K14846 ribosome
production factor 1
Length=389
Score = 31.6 bits (70), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 2 QPPRLIVFHLPHGPTAHFTLSGVCLRHDL 30
+P L + HLP GPT++F LS V L ++
Sbjct 246 RPYGLYIVHLPDGPTSYFRLSSVRLAQEM 274
> dre:564551 rexo1, fb77b11, si:dkey-45m5.5, wu:fb77b11; REX1,
RNA exonuclease 1 homolog (S. cerevisiae); K14570 RNA exonuclease
1 [EC:3.1.-.-]
Length=1207
Score = 30.0 bits (66), Expect = 2.3, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query 13 HG-PTAHFTLSGVCLRHDLPERAPNMSQVLFCCCCSA 48
HG P H +SG + H+LPE+ N CC C A
Sbjct 928 HGYPLPHPEISGHAVVHNLPEKKNNDPFSKICCRCGA 964
Lambda K H
0.330 0.141 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40