bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3953_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
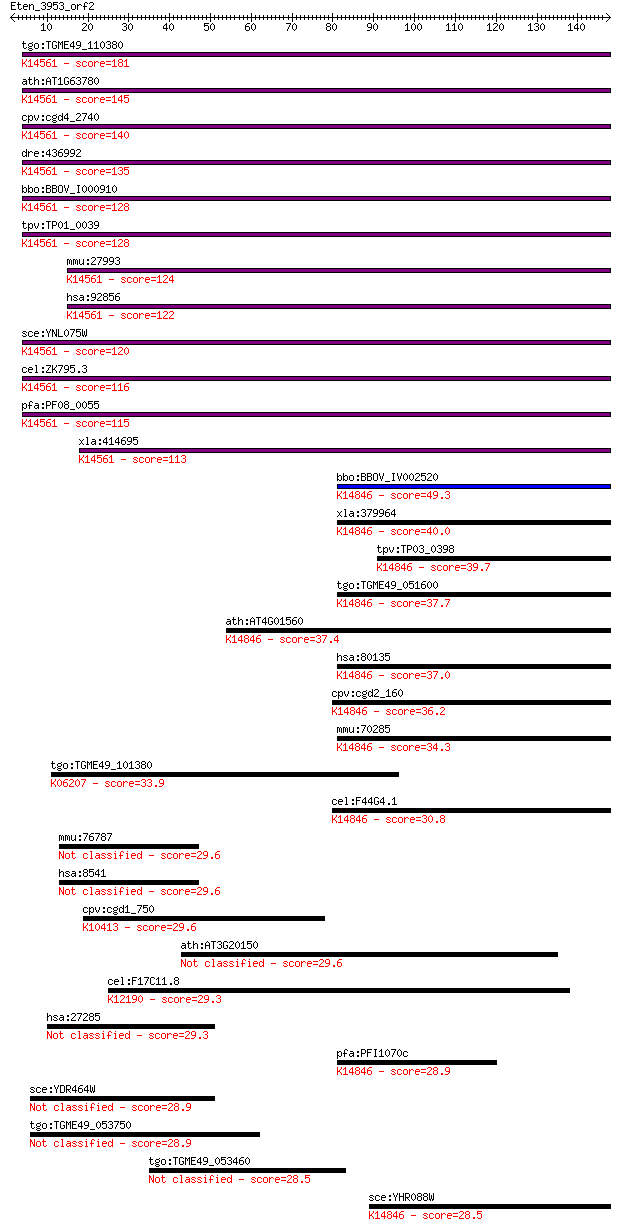
tgo:TGME49_110380 U3 small nucleolar ribonucleoprotein protein... 181 7e-46
ath:AT1G63780 IMP4; IMP4; K14561 U3 small nucleolar ribonucleo... 145 6e-35
cpv:cgd4_2740 IMP4 U3 small nucleolar ribonucleoprotein ; K145... 140 1e-33
dre:436992 imp4, zgc:86740; IMP4, U3 small nucleolar ribonucle... 135 4e-32
bbo:BBOV_I000910 16.m00780; Brix domain containing protein; K1... 128 5e-30
tpv:TP01_0039 U3 small nuclear ribonucleoprotein; K14561 U3 sm... 128 7e-30
mmu:27993 Imp4, AA409888, AV031295, D1Wsu40e; IMP4, U3 small n... 124 8e-29
hsa:92856 IMP4, BXDC4, MGC19606; IMP4, U3 small nucleolar ribo... 122 4e-28
sce:YNL075W IMP4; Component of the SSU processome, which is re... 120 1e-27
cel:ZK795.3 hypothetical protein; K14561 U3 small nucleolar ri... 116 3e-26
pfa:PF08_0055 U3 small nucleolar ribonucleoprotein protein, pu... 115 5e-26
xla:414695 imp4, MGC83136; IMP4, U3 small nucleolar ribonucleo... 113 2e-25
bbo:BBOV_IV002520 21.m02859; RNA processing factor 1; K14846 r... 49.3 5e-06
xla:379964 bxdc5, MGC53934, rpf1; RNA processing factor 1; K14... 40.0 0.003
tpv:TP03_0398 hypothetical protein; K14846 ribosome production... 39.7 0.003
tgo:TGME49_051600 RNA processing factor, putative ; K14846 rib... 37.7 0.013
ath:AT4G01560 MEE49; MEE49 (maternal effect embryo arrest 49);... 37.4 0.015
hsa:80135 RPF1, BXDC5, DKFZp761G0415, DKFZp761M0215, FLJ12475,... 37.0 0.023
cpv:cgd2_160 IMP4-like U3 small nucleolar ribonucleoprotein (s... 36.2 0.041
mmu:70285 Rpf1, 2210420E24Rik, 2310066N05Rik, Bxdc5, MGC102255... 34.3 0.15
tgo:TGME49_101380 GTP-binding protein TypA, putative (EC:2.7.7... 33.9 0.19
cel:F44G4.1 hypothetical protein; K14846 ribosome production f... 30.8 1.5
mmu:76787 Ppfia3, 2410127E16Rik; protein tyrosine phosphatase,... 29.6 3.3
hsa:8541 PPFIA3, KIAA0654, LPNA3, MGC126567, MGC126569; protei... 29.6 3.4
cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain 1,... 29.6 3.5
ath:AT3G20150 kinesin motor family protein 29.6 3.6
cel:F17C11.8 tag-318; Temporarily Assigned Gene name family me... 29.3 4.6
hsa:27285 TEKT2, TEKTB1, TEKTIN-T, h-tektin-t; tektin 2 (testi... 29.3 5.1
pfa:PFI1070c nucleolar preribosomal assembly protein, putative... 28.9 6.1
sce:YDR464W SPP41; Protein involved in negative regulation of ... 28.9 6.5
tgo:TGME49_053750 hypothetical protein 28.9 6.5
tgo:TGME49_053460 hypothetical protein 28.5 7.1
sce:YHR088W RPF1; Nucleolar protein involved in the assembly a... 28.5 8.3
> tgo:TGME49_110380 U3 small nucleolar ribonucleoprotein protein,
putative (EC:3.1.2.15); K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=314
Score = 181 bits (459), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 88/144 (61%), Positives = 113/144 (78%), Gaps = 4/144 (2%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLRRT RLRKEYLFRK+ EEK R LA+RK+++KE LE GK +P++LR A +L
Sbjct 1 MLRRTVRLRKEYLFRKATEEKARVLADRKKRLKEALERGKAVPSDLRGEAGQ----KLLP 56
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
LDL+D T TH+DDEYAYAGVEDP + LTTSR+PSSRLQQFVKEL LL+PN +RIN
Sbjct 57 TLDLEDVNTAQTTTHIDDEYAYAGVEDPKVVLTTSRNPSSRLQQFVKELRLLIPNSQRIN 116
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG V+++L++LCRS+ +TDL+++
Sbjct 117 RGGYVVNDLVELCRSNNVTDLIII 140
> ath:AT1G63780 IMP4; IMP4; K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=294
Score = 145 bits (365), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 71/144 (49%), Positives = 100/144 (69%), Gaps = 5/144 (3%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
M RR RL+KEY++RKSLE ER++ E+K+ ++E L+ GKP+PT LR A +L Q
Sbjct 1 MQRRLVRLKKEYIYRKSLEGDERKVYEQKRLIREALQEGKPIPTELRNVEA-----KLRQ 55
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
+DL+D T P++H+DDEYA A DP I LTTSR+PS+ L +F KEL + PN +RIN
Sbjct 56 EIDLEDQNTAVPRSHIDDEYANATEADPKILLTTSRNPSAPLIRFTKELKFVFPNSQRIN 115
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG+ V+ E+++ RS TD++LV
Sbjct 116 RGSQVISEIIETARSHDFTDVILV 139
> cpv:cgd4_2740 IMP4 U3 small nucleolar ribonucleoprotein ; K14561
U3 small nucleolar ribonucleoprotein protein IMP4
Length=288
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 73/144 (50%), Positives = 98/144 (68%), Gaps = 6/144 (4%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLRR AR RKEY+ RK LE E+ + ER + KE L+ G+ +PT + + A
Sbjct 1 MLRRNARQRKEYISRKGLERVEKNIFERSVRFKEALDEGRQIPTEYKNVSDKIVAN---- 56
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
+DL D ++ PK+ VDDEY+ +G+ +P I +TTSRSPSSRL QFVKELNL++PN RIN
Sbjct 57 -IDLFD-TSIDPKSIVDDEYSLSGLYEPKILITTSRSPSSRLLQFVKELNLVIPNSFRIN 114
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG VL++ LCRS+G TDL++V
Sbjct 115 RGGYVLNDFGDLCRSNGATDLVVV 138
> dre:436992 imp4, zgc:86740; IMP4, U3 small nucleolar ribonucleoprotein,
homolog (yeast); K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=291
Score = 135 bits (340), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 65/144 (45%), Positives = 96/144 (66%), Gaps = 5/144 (3%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLRR RLRKEYL+RK+ E++ R + E+KQ++K L+ + LPT +R A + +
Sbjct 1 MLRREVRLRKEYLYRKAQEDRLRTIEEKKQKLKSALDENRLLPTEVRKEALE-----VQK 55
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
+L+ DD +H+DDEY +AGVEDP I +TTSR PSSRL+ F KE+ L+ P +R+N
Sbjct 56 LLEFDDEGGEGVSSHMDDEYKWAGVEDPKIMVTTSRDPSSRLKMFAKEMKLMFPGAQRMN 115
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG + L+ C+S+ +TDL++V
Sbjct 116 RGNHEVKTLVHACKSNNVTDLVIV 139
> bbo:BBOV_I000910 16.m00780; Brix domain containing protein;
K14561 U3 small nucleolar ribonucleoprotein protein IMP4
Length=293
Score = 128 bits (322), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 62/144 (43%), Positives = 93/144 (64%), Gaps = 5/144 (3%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLR+T R R+EYLF K+ EE++ + +++KE L+ KP+PT LR + +
Sbjct 1 MLRKTLRERREYLFAKAQEERQATSLKNARKLKEALKDNKPIPTELRNLSGTVHDS---- 56
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
LDL D TH DDEYA+ G++ P I +TTSR PSSRL QF KE+ L++PN R+N
Sbjct 57 -LDLLDTHHREEHTHADDEYAFVGIKTPKIMVTTSRDPSSRLSQFAKEVRLIIPNSERMN 115
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG+ +L +L+ C+S ++D++L+
Sbjct 116 RGSYILKDLVTFCKSKDVSDIVLL 139
> tpv:TP01_0039 U3 small nuclear ribonucleoprotein; K14561 U3
small nucleolar ribonucleoprotein protein IMP4
Length=302
Score = 128 bits (321), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 64/154 (41%), Positives = 99/154 (64%), Gaps = 15/154 (9%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLR+T R R+EY+F KS E K++ ++ +++ E L+ KP+PT LR + A+
Sbjct 1 MLRKTLRQRREYIFLKSRENKDKYSLKKARELNEALKNNKPIPTELRNQSGVVQAS---- 56
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGV----------EDPSIALTTSRSPSSRLQQFVKELN 113
LDL D + +HVD+EYA+ G+ +DP I +TTSR+PSSRL QF KEL
Sbjct 57 -LDLLDAKRREEYSHVDNEYAFCGMLHYFTKNLGFKDPKILITTSRNPSSRLTQFAKELC 115
Query 114 LLLPNCRRINRGTIVLDELLQLCRSSGITDLLLV 147
L++PN R+NRG+ +L +L++ CRS ++D++L+
Sbjct 116 LIIPNSERLNRGSYILKDLVEFCRSKDVSDMVLI 149
> mmu:27993 Imp4, AA409888, AV031295, D1Wsu40e; IMP4, U3 small
nucleolar ribonucleoprotein, homolog (yeast); K14561 U3 small
nucleolar ribonucleoprotein protein IMP4
Length=291
Score = 124 bits (312), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 62/133 (46%), Positives = 88/133 (66%), Gaps = 5/133 (3%)
Query 15 YLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQVLDLDDPQTLA 74
YL+RK+ EE +R + E+K++VK LE + +PT LR A A L L+ DD
Sbjct 12 YLYRKAREEAQRSVQEKKERVKRALEENQLIPTELRREALA-----LQGSLEFDDAGGEG 66
Query 75 PKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQ 134
+HVDDEY +AGVEDP + +TTSR PSSRL+ F KEL L+ P +R+NRG + L++
Sbjct 67 VTSHVDDEYRWAGVEDPKVMITTSRDPSSRLKMFAKELKLVFPGAQRMNRGRHEVGALVR 126
Query 135 LCRSSGITDLLLV 147
C+++G+TDLL+V
Sbjct 127 ACKANGVTDLLVV 139
> hsa:92856 IMP4, BXDC4, MGC19606; IMP4, U3 small nucleolar ribonucleoprotein,
homolog (yeast); K14561 U3 small nucleolar
ribonucleoprotein protein IMP4
Length=291
Score = 122 bits (306), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 61/133 (45%), Positives = 87/133 (65%), Gaps = 5/133 (3%)
Query 15 YLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQVLDLDDPQTLA 74
YL+RK+ EE +R ERK++++ LE + +PT LR A A L L+ DD
Sbjct 12 YLYRKAREEAQRSAQERKERLRRALEENRLIPTELRREALA-----LQGSLEFDDAGGEG 66
Query 75 PKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQ 134
+HVDDEY +AGVEDP + +TTSR PSSRL+ F KEL L+ P +R+NRG + L++
Sbjct 67 VTSHVDDEYRWAGVEDPKVMITTSRDPSSRLKMFAKELKLVFPGAQRMNRGRHEVGALVR 126
Query 135 LCRSSGITDLLLV 147
C+++G+TDLL+V
Sbjct 127 ACKANGVTDLLVV 139
> sce:YNL075W IMP4; Component of the SSU processome, which is
required for pre-18S rRNA processing; interacts with Mpp10p;
member of a superfamily of proteins that contain a sigma(70)-like
motif and associate with RNAs; K14561 U3 small nucleolar
ribonucleoprotein protein IMP4
Length=290
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 68/149 (45%), Positives = 92/149 (61%), Gaps = 12/149 (8%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLRR AR R+EYL+RK+ E ++ QL +++Q +K+ L GKPLP L + R Q
Sbjct 1 MLRRQARERREYLYRKAQELQDSQLQQKRQIIKQALAQGKPLPKELAEDESLQKDFRYDQ 60
Query 64 VL----DLDDPQTLAPKTHVDDEYA-YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPN 118
L + DD Q VDDEYA +G+ DP I +TTSR PS+RL QF KE+ LL PN
Sbjct 61 SLKESEEADDLQ-------VDDEYAATSGIMDPRIIVTTSRDPSTRLSQFAKEIKLLFPN 113
Query 119 CRRINRGTIVLDELLQLCRSSGITDLLLV 147
R+NRG V+ L+ C+ SG TDL+++
Sbjct 114 AVRLNRGNYVMPNLVDACKKSGTTDLVVL 142
> cel:ZK795.3 hypothetical protein; K14561 U3 small nucleolar
ribonucleoprotein protein IMP4
Length=292
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 59/144 (40%), Positives = 88/144 (61%), Gaps = 10/144 (6%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
M+RR RLR+E++FRKSLEEK++ L E++++++ LE + NLR A LA+
Sbjct 1 MIRRENRLRREFIFRKSLEEKQKSLEEKREKIRNALENNTKIDYNLRKDAI-----ELAK 55
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
D Q D EY +AG +DP I +TTSR PSSRL+ F KE+ L+ PN +RIN
Sbjct 56 GSDWGGQQY-----ETDSEYRWAGAQDPKIVITTSRDPSSRLKMFAKEMKLIFPNAQRIN 110
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG + +++Q ++ TDL++
Sbjct 111 RGHYDVKQVVQASKAQDSTDLIIF 134
> pfa:PF08_0055 U3 small nucleolar ribonucleoprotein protein,
putative; K14561 U3 small nucleolar ribonucleoprotein protein
IMP4
Length=362
Score = 115 bits (288), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 55/144 (38%), Positives = 95/144 (65%), Gaps = 7/144 (4%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLRR RLRKEYL+ K +E+++++ AE+ + +KE+ + K + +L+ + L +
Sbjct 1 MLRRNIRLRKEYLYLKKVEDEKKKYAEKIKSIKESYDKNKKIRGDLKDEESE-----LRK 55
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
++L D ++ K VDDEY + G+E+P + +TTSR+PSS L+ F KEL L++PN +IN
Sbjct 56 NMNLYDEKSFDRK--VDDEYFFCGLENPRVLITTSRNPSSTLENFAKELKLIIPNSEKIN 113
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG+ + ++L R + ITD++++
Sbjct 114 RGSYFIKDILNFARKNNITDVIIL 137
> xla:414695 imp4, MGC83136; IMP4, U3 small nucleolar ribonucleoprotein,
homolog; K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=291
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 57/130 (43%), Positives = 83/130 (63%), Gaps = 5/130 (3%)
Query 18 RKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQVLDLDDPQTLAPKT 77
+K+ E K + E+K+++K LE + +PT +R A L + L+ DD +
Sbjct 15 KKAQEAKLHSIEEKKERLKRALEENRLIPTEIRREALT-----LQKQLEFDDEGGEGVSS 69
Query 78 HVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCR 137
HVDDEY +AGVEDP I +TTSR PSSRL+ F KE+ L+ PN +R+NRG + L+Q CR
Sbjct 70 HVDDEYKWAGVEDPKIMITTSRDPSSRLKIFAKEMRLIFPNAQRMNRGKHEVGALVQACR 129
Query 138 SSGITDLLLV 147
++ +TDLL+V
Sbjct 130 ANDVTDLLIV 139
> bbo:BBOV_IV002520 21.m02859; RNA processing factor 1; K14846
ribosome production factor 1
Length=301
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 43/69 (62%), Gaps = 2/69 (2%)
Query 81 DEYA--YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRS 138
DE+A + G +P + LT+SR PS ++++F+KEL L++PN R L+++++
Sbjct 83 DEFAAHFRGDREPKLYLTSSRRPSEKMRRFIKELLLIIPNIYYFARDENKLEDMVRHATD 142
Query 139 SGITDLLLV 147
G + +LL+
Sbjct 143 GGFSAILLI 151
> xla:379964 bxdc5, MGC53934, rpf1; RNA processing factor 1; K14846
ribosome production factor 1
Length=343
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 37/69 (53%), Gaps = 2/69 (2%)
Query 81 DEYA--YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRS 138
DE+A + P I +TTS P R +F ++L+ ++PN R + L +++ C S
Sbjct 124 DEFAPYFNKQTTPKILITTSDRPRGRSVRFTEQLSSIIPNSDVYYRRGLALKKIIPQCVS 183
Query 139 SGITDLLLV 147
TDLL++
Sbjct 184 RDYTDLLVI 192
> tpv:TP03_0398 hypothetical protein; K14846 ribosome production
factor 1
Length=321
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 91 PSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRSSGITDLLLV 147
P + +T++R PS +++ ++KEL L+L N R L E+++ +G T +LLV
Sbjct 115 PKLLITSTRRPSEKMRTYMKELLLVLSNSYYFAREEYKLKEVVKHATENGFTSILLV 171
> tgo:TGME49_051600 RNA processing factor, putative ; K14846 ribosome
production factor 1
Length=389
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 35/69 (50%), Gaps = 2/69 (2%)
Query 81 DEYA-YAGVE-DPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRS 138
DE+A Y E P + +TT++ ++ + F+KE+ + PN RG + + +
Sbjct 172 DEFAAYFNREVTPKLMITTNKRVTADMYSFLKEVLYIFPNAFFYKRGPYQIKNICKFAVK 231
Query 139 SGITDLLLV 147
G TDLL+
Sbjct 232 QGFTDLLVF 240
> ath:AT4G01560 MEE49; MEE49 (maternal effect embryo arrest 49);
K14846 ribosome production factor 1
Length=343
Score = 37.4 bits (85), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 49/118 (41%), Gaps = 24/118 (20%)
Query 54 AAAAAARLAQVLDLDDPQTLAPKT------------HVDDEYAYAGVED----------- 90
A AA + A L + PQ + PKT DDE +A ++
Sbjct 66 ARDAAEKRALELGEEPPQKMIPKTIENTRESDETVCRPDDEELFADIDADEFNPVLRREI 125
Query 91 -PSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRSSGITDLLLV 147
P + LTT R S+R + EL ++PN RGT L ++++ T L++V
Sbjct 126 APKVLLTTCRFNSTRGPALISELLSVIPNSHYQKRGTYDLKKIVEYATKKDFTSLIVV 183
> hsa:80135 RPF1, BXDC5, DKFZp761G0415, DKFZp761M0215, FLJ12475,
RP11-118B23.1; ribosome production factor 1 homolog (S. cerevisiae);
K14846 ribosome production factor 1
Length=349
Score = 37.0 bits (84), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 81 DEYA--YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRS 138
DE+A + P I +TTS P R + ++L+ ++PN R + L +++ C +
Sbjct 130 DEFASYFNKQTSPKILITTSDRPHGRTVRLCEQLSTVIPNSHVYYRRGLALKKIIPQCIA 189
Query 139 SGITDLLLV 147
TDL+++
Sbjct 190 RDFTDLIVI 198
> cpv:cgd2_160 IMP4-like U3 small nucleolar ribonucleoprotein
(snoRNP) ; K14846 ribosome production factor 1
Length=359
Score = 36.2 bits (82), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 17/74 (22%), Positives = 37/74 (50%), Gaps = 6/74 (8%)
Query 80 DDEYA--YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCR 137
+DE++ ++ P + +TT+ S + F+KE+ ++PNC R + ++++
Sbjct 119 NDEFSDYFSRKRSPKLLITTNNKASKHMYNFLKEIVSIIPNCDFYKRNGFNIKDIIKEVN 178
Query 138 SSG----ITDLLLV 147
G TD+L+
Sbjct 179 EKGTENNYTDILIF 192
> mmu:70285 Rpf1, 2210420E24Rik, 2310066N05Rik, Bxdc5, MGC102255;
ribosome production factor 1 homolog (S. cerevisiae); K14846
ribosome production factor 1
Length=212
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 81 DEYA--YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRS 138
DE+A + P I +TTS P R + ++L+ ++P+ R + L +++ C +
Sbjct 130 DEFASYFNRQTSPKILITTSDRPHGRTVRLCEQLSTVIPDSHVYYRRGLALKKIIPQCIA 189
Query 139 SGITDLLLV 147
TDL+++
Sbjct 190 RDFTDLIVI 198
> tgo:TGME49_101380 GTP-binding protein TypA, putative (EC:2.7.7.4);
K06207 GTP-binding protein
Length=749
Score = 33.9 bits (76), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 43/86 (50%), Gaps = 12/86 (13%)
Query 11 LRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLA-QVLDLDD 69
LRKE + + EE++++ R + + + + LP ++ A + A R+ V+DL D
Sbjct 310 LRKEGVAERGPEEEQKEAFPRSENLA-VVRSASSLPGDILVLAGVSTAVRVGDSVVDLVD 368
Query 70 PQTLAPKTHVDDEYAYAGVEDPSIAL 95
P+ LAP V DPS+++
Sbjct 369 PRPLAPMK----------VADPSVSI 384
> cel:F44G4.1 hypothetical protein; K14846 ribosome production
factor 1
Length=384
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 80 DDEYAYAGVEDPSIALTTSRSPSSRLQ--QFVKELNLLLPNCRRINRGTIVLDELLQLCR 137
+DE+A + S + + +P +++ +F EL +PN R ++L +++ +
Sbjct 164 NDEFAPYFNRETSPKVMITMTPKAKITTFKFCFELQKCIPNSEIFTRKNVLLKTIIEQAK 223
Query 138 SSGITDLLLV 147
TDLL+V
Sbjct 224 EREFTDLLVV 233
> mmu:76787 Ppfia3, 2410127E16Rik; protein tyrosine phosphatase,
receptor type, f polypeptide (PTPRF), interacting protein
(liprin), alpha 3
Length=1194
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 27/38 (71%), Gaps = 5/38 (13%)
Query 13 KEYLFRKSLEEKERQLAER----KQQVKETLETGKPLP 46
KE L+R+S EEK RQLAE KQ++++TL+ + LP
Sbjct 336 KESLYRQS-EEKSRQLAEWLDDAKQKLQQTLQKAETLP 372
> hsa:8541 PPFIA3, KIAA0654, LPNA3, MGC126567, MGC126569; protein
tyrosine phosphatase, receptor type, f polypeptide (PTPRF),
interacting protein (liprin), alpha 3
Length=1194
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 27/38 (71%), Gaps = 5/38 (13%)
Query 13 KEYLFRKSLEEKERQLAER----KQQVKETLETGKPLP 46
KE L+R+S EEK RQLAE KQ++++TL+ + LP
Sbjct 336 KESLYRQS-EEKSRQLAEWLDDAKQKLQQTLQKAETLP 372
> cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain
1, cytosolic
Length=5246
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 19 KSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQVLDLDDPQTLAPKT 77
KSL+E+++ +AER +V+ L+ +P+ A + L ++ + +P L KT
Sbjct 3643 KSLDEQQKVIAERSSEVEIQLKDVEPILREAENAVSNIPKKNLDELRSMANPPGLVKKT 3701
> ath:AT3G20150 kinesin motor family protein
Length=1114
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 21/93 (22%), Positives = 45/93 (48%), Gaps = 6/93 (6%)
Query 43 KPLPTNLRAAAAAAAAARLAQVLDLDDPQTLAPKTHVD-DEYAYAGVEDPSIALTTSRSP 101
K P N R + ++ + +DD + + + V+ +++ + E S A S
Sbjct 540 KKFPVN-RDSVNSSFVTAFGESELMDDDEICSEEVEVEENDFGESLEEHDSAATVCKSSE 598
Query 102 SSRLQQFVKELNLLLPNCRRINRGTIVLDELLQ 134
SR+++FV E ++ + CR+ +++L E +Q
Sbjct 599 KSRIEEFVSENSISISPCRQ----SLILQEPIQ 627
> cel:F17C11.8 tag-318; Temporarily Assigned Gene name family
member (tag-318); K12190 ESCRT-II complex subunit VPS36
Length=383
Score = 29.3 bits (64), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 56/120 (46%), Gaps = 13/120 (10%)
Query 25 ERQLAERKQQVKETL-----ETGKPLPT--NLRAAAAAAAAARLAQVLDLDDPQTLAPKT 77
ER+LAE Q+ ET+ + K + T + A + + + ++ ++ + +T+A K+
Sbjct 158 ERRLAENHQKTHETITQAFDDMSKLMETAREMVALSKSISEKVRSRKGEISEDETIAFKS 217
Query 78 HVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCR 137
++ GV DP T S S Q+ KE++ +L + N G L E+ CR
Sbjct 218 YL----LSLGVSDPVTKSTFVGSDSEYFQKLAKEISDVLYEHIKENGGMCALPEV--YCR 271
> hsa:27285 TEKT2, TEKTB1, TEKTIN-T, h-tektin-t; tektin 2 (testicular)
Length=430
Score = 29.3 bits (64), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 10 RLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLR 50
R+ E+ FRK L E E+ +E K Q K TLE L ++R
Sbjct 258 RVATEFAFRKRLREMEKVYSELKWQEKNTLEEIAELQEDIR 298
> pfa:PFI1070c nucleolar preribosomal assembly protein, putative;
K14846 ribosome production factor 1
Length=328
Score = 28.9 bits (63), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Query 81 DEYA--YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNC 119
DE++ + ++P+I +T+ + PS F+KEL+L P+
Sbjct 110 DEFSDYFKNQKEPNIIITSIKRPSKNTILFMKELSLTFPHI 150
> sce:YDR464W SPP41; Protein involved in negative regulation of
expression of spliceosome components PRP4 and PRP3
Length=1435
Score = 28.9 bits (63), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 31/45 (68%), Gaps = 4/45 (8%)
Query 6 RRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLR 50
R+TAR +E +K LE+ ++LAE ++++K+ +E G P P +LR
Sbjct 622 RQTAR--EERRHKKKLEK--QRLAEEQEELKKIVERGPPYPPDLR 662
> tgo:TGME49_053750 hypothetical protein
Length=8002
Score = 28.9 bits (63), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 28/56 (50%), Gaps = 0/56 (0%)
Query 6 RRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARL 61
+R+A +F K EE+E + + Q ET + GKP+P AA A+ ++
Sbjct 6120 KRSAEPGSWSVFSKHGEEQETRYSAAPQTNAETDDQGKPVPNESGLAAPDASDVKM 6175
> tgo:TGME49_053460 hypothetical protein
Length=193
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 35 VKETLETGKPLPTNLRAAAAAAAAARLAQVLDLDDPQ-TLAPKTHVDDE 82
V E+LET + T L A+ AAA V ++ PQ T PK H+DD+
Sbjct 124 VTESLETAERTLTELPTASEAAATTGEVPVNEMWAPQATELPKQHIDDD 172
> sce:YHR088W RPF1; Nucleolar protein involved in the assembly
and export of the large ribosomal subunit; constituent of 66S
pre-ribosomal particles; contains a sigma(70)-like motif,
which is thought to bind RNA; K14846 ribosome production factor
1
Length=295
Score = 28.5 bits (62), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Query 89 EDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRG-TIVLDELLQLCRSSGITDLLLV 147
E P I LTT+ + +F L +LPN + R L E+ +C TD++++
Sbjct 91 EPPKIFLTTNVNAKKSAYEFANILIEILPNVTFVKRKFGYKLKEISDICIKRNFTDIVII 150
Lambda K H
0.317 0.132 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40