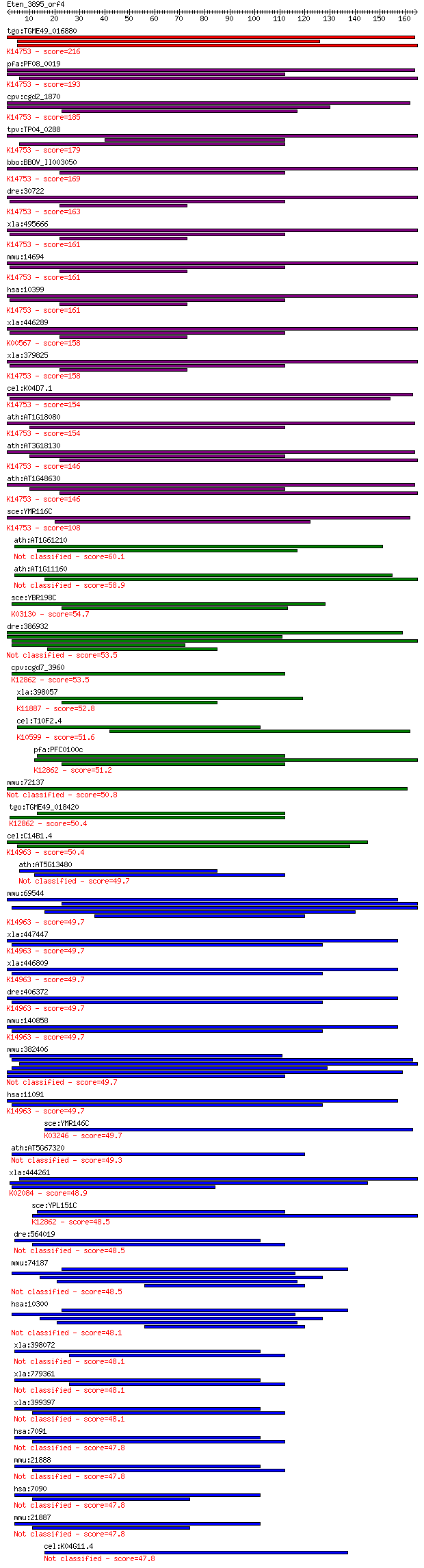
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3895_orf4
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_016880 receptor for activated C kinase, RACK protei... 216 2e-56
pfa:PF08_0019 PfRACK; receptor for activated C kinase homolog,... 193 3e-49
cpv:cgd2_1870 guanine nucleotide-binding protein ; K14753 guan... 185 5e-47
tpv:TP04_0288 guanine nucleotide-binding protein; K14753 guani... 179 3e-45
bbo:BBOV_II003050 18.m06254; receptor for activated C kinase, ... 169 3e-42
dre:30722 gnb2l1, rack1, wu:fb80d08, wu:fk65d12; guanine nucle... 163 3e-40
xla:495666 gnb2l1, gnb2-rs1, h12.3, hlc-7, pig21, rack1; guani... 161 1e-39
mmu:14694 Gnb2l1, AL033335, GB-like, Gnb2-rs1, Rack1, p205; gu... 161 1e-39
hsa:10399 GNB2L1, Gnb2-rs1, H12.3, HLC-7, PIG21, RACK1; guanin... 161 1e-39
xla:446289 MGC130692; hypothetical protein LOC446289; K00567 m... 158 9e-39
xla:379825 gnb2l1, MGC53289; guanine nucleotide binding protei... 158 9e-39
cel:K04D7.1 rack-1; RACK1 (mammalian Receptor of Activated C K... 154 1e-37
ath:AT1G18080 ATARCA; ATARCA; nucleotide binding; K14753 guani... 154 1e-37
ath:AT3G18130 RACK1C_AT (RECEPTOR FOR ACTIVATED C KINASE 1 C);... 146 3e-35
ath:AT1G48630 RACK1B_AT (RECEPTOR FOR ACTIVATED C KINASE 1 B);... 146 3e-35
sce:YMR116C ASC1, CPC2; Asc1p; K14753 guanine nucleotide-bindi... 108 7e-24
ath:AT1G61210 WD-40 repeat family protein / katanin p80 subuni... 60.1 3e-09
ath:AT1G11160 nucleotide binding 58.9 8e-09
sce:YBR198C TAF5, TAF90; Taf5p; K03130 transcription initiatio... 54.7 1e-07
dre:386932 poc1b, TUWD12, fl36w17, wdr51b, wu:fl36w17, zgc:635... 53.5 3e-07
cpv:cgd7_3960 pleiotropic regulator 1 ; K12862 pleiotropic reg... 53.5 3e-07
xla:398057 paaf1, paaf, rpn14, wdr71; proteasomal ATPase-assoc... 52.8 5e-07
cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing f... 51.6 1e-06
pfa:PFC0100c regulatory protein, putative; K12862 pleiotropic ... 51.2 1e-06
mmu:72137 Wdsub1, 1700048E19Rik, 2610014F08Rik; WD repeat, SAM... 50.8 2e-06
tgo:TGME49_018420 pleiotropic regulator 1, putative (EC:2.7.11... 50.4 2e-06
cel:C14B1.4 tag-125; Temporarily Assigned Gene name family mem... 50.4 3e-06
ath:AT5G13480 FY; FY; protein binding 49.7 4e-06
mmu:69544 Wdr5b, 2310009C03Rik, AI606931; WD repeat domain 5B;... 49.7 4e-06
xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repe... 49.7 4e-06
xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat dom... 49.7 5e-06
dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat d... 49.7 5e-06
mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3... 49.7 5e-06
mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar prote... 49.7 5e-06
hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPAS... 49.7 5e-06
sce:YMR146C TIF34; Tif34p; K03246 translation initiation facto... 49.7 5e-06
ath:AT5G67320 HOS15; HOS15 (high expression of osmotically res... 49.3 6e-06
xla:444261 apaf1, MGC80868, apaf-1; apoptotic peptidase activa... 48.9 7e-06
sce:YPL151C PRP46, NTC50; Member of the NineTeen Complex (NTC)... 48.5 1e-05
dre:564019 tle1; transducin-like enhancer of split 1 (E(sp1) h... 48.5 1e-05
mmu:74187 Katnb1, 2410003J24Rik, KAT; katanin p80 (WD40-contai... 48.5 1e-05
hsa:10300 KATNB1, KAT; katanin p80 (WD repeat containing) subu... 48.1 1e-05
xla:398072 tle2, esg, esg1, esg2, esg3, grg2, grg3, tle3, tle3... 48.1 1e-05
xla:779361 tle1, esg, esg1, grg1, tle3b, xgrg1; transducin-lik... 48.1 1e-05
xla:399397 tle4, ESG, MGC86378, XGrg-4, XTCF-4, Xgrg4, Xtle4, ... 48.1 1e-05
hsa:7091 TLE4, BCE-1, BCE1, E(spI), ESG, ESG4, GRG4; transduci... 47.8 1e-05
mmu:21888 Tle4, 5730411M05Rik, AA792082, Bce-1, Bce1, ESTM13, ... 47.8 2e-05
hsa:7090 TLE3, ESG, ESG3, FLJ39460, GRG3, HsT18976, KIAA1547; ... 47.8 2e-05
mmu:21887 Tle3, 2610103N05Rik, ESG, Grg3a, Grg3b, mKIAA1547; t... 47.8 2e-05
cel:K04G11.4 hypothetical protein 47.8 2e-05
> tgo:TGME49_016880 receptor for activated C kinase, RACK protein,
putative (EC:2.7.11.7); K14753 guanine nucleotide-binding
protein subunit beta-2-like 1 protein
Length=321
Score = 216 bits (551), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 100/163 (61%), Positives = 120/163 (73%), Gaps = 0/163 (0%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
DWVSCVRF P +PLI CGWDKLVKVW+L+ CKL NLVG T VLYTVT+SPDGSLCA
Sbjct 154 DWVSCVRFSPSANKPLIVSCGWDKLVKVWNLSNCKLRTNLVGHTSVLYTVTISPDGSLCA 213
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDGV +LWD++EGKHL+SLD + IN+L F PC YW+ ATDK KIW E K VL
Sbjct 214 SGGKDGVAMLWDVNEGKHLYSLDSNSTINALCFSPCNYWLCAATDKSVKIWDLENKNVLS 273
Query 121 DISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAE 163
+I+P+ + G PW NWS++GR LF+G+ G I VYEV+
Sbjct 274 EITPEKTNRSGAPWCTSLNWSHDGRTLFVGTFTGAINVYEVSS 316
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 38/136 (27%), Positives = 55/136 (40%), Gaps = 15/136 (11%)
Query 5 CVRFFPFP--KEPLIFLCGWDKLVKVWSL--------TTCKLPANLVGQTFVLYTVTVSP 54
CV P K I D V VW+L + L G + + V ++
Sbjct 19 CVTAISTPSLKSNTIVSSSRDHKVIVWTLNDNPDDSGSVGYARRALTGHSQCVQDVVINS 78
Query 55 DGSLCAFGGKDGVTILWDLSEGKHLFSLDG-SCPINSLGFFP-CKYWVGGATDKFFKIW- 111
DG G D LWDL+ G + S G + +NS+ F P + V G+ D+ K+W
Sbjct 79 DGQFALSGSWDKTLRLWDLNAGVTVRSFQGHTSDVNSVAFSPDNRQIVSGSRDRTIKLWN 138
Query 112 --APEKKKVLDDISPD 125
A K ++DD D
Sbjct 139 TLAECKYTIVDDQHND 154
Score = 35.4 bits (80), Expect = 0.087, Method: Compositional matrix adjust.
Identities = 39/166 (23%), Positives = 63/166 (37%), Gaps = 12/166 (7%)
Query 5 CVRFFPFPKEPLIFLCG-WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
CV+ + L G WDK +++W L + G T + +V SPD G
Sbjct 70 CVQDVVINSDGQFALSGSWDKTLRLWDLNAGVTVRSFQGHTSDVNSVAFSPDNRQIVSGS 129
Query 64 KDGVTILWD-LSEGKHLFSLDGSCP-INSLGFFPCK---YWVGGATDKFFKIWAPEKKKV 118
+D LW+ L+E K+ D ++ + F P V DK K+W K+
Sbjct 130 RDRTIKLWNTLAECKYTIVDDQHNDWVSCVRFSPSANKPLIVSCGWDKLVKVWNLSNCKL 189
Query 119 LDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ V + P +G G G +++V EG
Sbjct 190 RTNLVGHTSVLYTVTISP------DGSLCASGGKDGVAMLWDVNEG 229
> pfa:PF08_0019 PfRACK; receptor for activated C kinase homolog,
PfRACK; K14753 guanine nucleotide-binding protein subunit
beta-2-like 1 protein
Length=323
Score = 193 bits (490), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 91/163 (55%), Positives = 110/163 (67%), Gaps = 0/163 (0%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
DW++CVRF P P + +I CGWDKLVKVW+L C L NL G T VL TVT+SPDGSLCA
Sbjct 158 DWITCVRFSPSPNQAIIVSCGWDKLVKVWNLKNCDLNKNLEGHTGVLNTVTISPDGSLCA 217
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDGV LWD+ EGKHL+SL+ INSL F PC YW+ ATD+F +IW E K ++
Sbjct 218 SGGKDGVAKLWDVKEGKHLYSLETGSTINSLCFSPCDYWLCAATDRFIRIWNLESKLIIS 277
Query 121 DISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAE 163
+I P K G+PW WS G+ L+ GS GNI VYEV +
Sbjct 278 EIYPVKQSKIGVPWCTSLTWSANGQLLYCGSTDGNIYVYEVKK 320
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 53/120 (44%), Gaps = 9/120 (7%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTT-------CKLPANLVGQTFVLYTVTVS 53
DWV+ V PK I DK + VW++ T +L G + + V++S
Sbjct 22 DWVTSVSTPTDPKLKTIVSASRDKKLIVWNINTDDDSGEIGTAKKSLTGHSQAINDVSIS 81
Query 54 PDGSLCAFGGKDGVTILWDLSEGKHLFSLDG-SCPINSLGFFP-CKYWVGGATDKFFKIW 111
DG G D LWDLS G+ + S G + + S+ F P + V + DK K+W
Sbjct 82 SDGLFALSGSWDHSVRLWDLSLGETIRSFIGHTSDVFSVSFSPDNRQIVSASRDKTIKLW 141
Score = 34.3 bits (77), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 35/165 (21%), Positives = 64/165 (38%), Gaps = 12/165 (7%)
Query 6 VRFFPFPKEPLIFLCG-WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
+ + L L G WD V++W L+ + + +G T +++V+ SPD +
Sbjct 75 INDVSISSDGLFALSGSWDHSVRLWDLSLGETIRSFIGHTSDVFSVSFSPDNRQIVSASR 134
Query 65 DGVTILWD-LSEGKHLFSLDGSCP-INSLGFFPC---KYWVGGATDKFFKIWAPEKKKVL 119
D LW+ L++ K+ + I + F P V DK K+W + +
Sbjct 135 DKTIKLWNTLAQCKYTITDQQHTDWITCVRFSPSPNQAIIVSCGWDKLVKVWNLKNCDLN 194
Query 120 DDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ V + P +G G G +++V EG
Sbjct 195 KNLEGHTGVLNTVTISP------DGSLCASGGKDGVAKLWDVKEG 233
> cpv:cgd2_1870 guanine nucleotide-binding protein ; K14753 guanine
nucleotide-binding protein subunit beta-2-like 1 protein
Length=313
Score = 185 bits (470), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 91/161 (56%), Positives = 111/161 (68%), Gaps = 1/161 (0%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
DWVSCVRF P +PL CGWDK+VKVWS K NLVG + VLYTVT++PDGSLCA
Sbjct 152 DWVSCVRFSPSTSQPLFVSCGWDKIVKVWSHEF-KPTCNLVGHSSVLYTVTIAPDGSLCA 210
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDGV +LWD+SEGKHL+SLD + IN+L F PC YW+ ATDK +IW E K +
Sbjct 211 SGGKDGVVMLWDVSEGKHLYSLDANHSINALCFSPCNYWLCAATDKCIRIWELEHKLPVA 270
Query 121 DISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEV 161
+ +P P+K G+PW WS +G+ LF GS NI VY+V
Sbjct 271 EFAPPAPIKNGLPWCVSLTWSNDGKTLFAGSTDSNIYVYKV 311
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 33/137 (24%), Positives = 54/137 (39%), Gaps = 27/137 (19%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCK--------LPANLVGQTFVLYTVTV 52
DWV+ + P ++ DK V VW+L+ L G + V V
Sbjct 16 DWVTSIAAPP-DNSDIVVSASRDKSVLVWNLSDANKDQHSIGSAKTRLTGHNQAVNDVAV 74
Query 53 SPDGSLCAFGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWA 112
S DGS G D L+D++ GK + + +VG +D F +
Sbjct 75 SSDGSYVVSGSCDKTLRLFDVNAGKSV-----------------RNFVGHTSDVFSVALS 117
Query 113 PEKKKVLDDISPDHPVK 129
P+ ++++ S DH +K
Sbjct 118 PDNRQIISG-SRDHTIK 133
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 47/109 (43%), Gaps = 25/109 (22%)
Query 23 DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD--------LS 74
DK ++++ + K N VG T +++V +SPD G +D +W+ L
Sbjct 87 DKTLRLFDVNAGKSVRNFVGHTSDVFSVALSPDNRQIISGSRDHTIKVWNAMGVCMYTLL 146
Query 75 EGKH-------LFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKK 116
+G+H FS S P+ F C + DK K+W+ E K
Sbjct 147 DGQHNDWVSCVRFSPSTSQPL----FVSCGW------DKIVKVWSHEFK 185
> tpv:TP04_0288 guanine nucleotide-binding protein; K14753 guanine
nucleotide-binding protein subunit beta-2-like 1 protein
Length=331
Score = 179 bits (455), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 87/164 (53%), Positives = 105/164 (64%), Gaps = 0/164 (0%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
DWVSCVRF P KEP+ GWDKL+KVW L TC+L L G V+Y+V++SPDGSLCA
Sbjct 162 DWVSCVRFSPSGKEPIFVSGGWDKLIKVWDLRTCQLKHTLYGHEGVVYSVSISPDGSLCA 221
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDGV LWD+ E L LD IN+L F PC YW+ ATDK KIW E K +L
Sbjct 222 SGGKDGVARLWDMKEANSLHLLDAGSTINALCFSPCNYWLCAATDKAIKIWDLENKNILA 281
Query 121 DISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
+I+ D K G+PW WS +GR L+ GS GNI VY+V+
Sbjct 282 EINNDRTKKVGLPWCVSLCWSPDGRVLYAGSTDGNIYVYQVSRN 325
Score = 37.7 bits (86), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 2/74 (2%)
Query 40 LVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CK 97
L G + + V++S DG G DG LWDL + + +G +NS+ F P +
Sbjct 72 LTGHSQTVQDVSMSSDGLFALSGSWDGTLRLWDLVKACSVRVFNGHTKDVNSVAFSPDNR 131
Query 98 YWVGGATDKFFKIW 111
+ G+ DK K+W
Sbjct 132 QIISGSRDKTIKLW 145
Score = 32.7 bits (73), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 46/112 (41%), Gaps = 6/112 (5%)
Query 6 VRFFPFPKEPLIFLCG-WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
V+ + L L G WD +++W L G T + +V SPD G +
Sbjct 79 VQDVSMSSDGLFALSGSWDGTLRLWDLVKACSVRVFNGHTKDVNSVAFSPDNRQIISGSR 138
Query 65 DGVTILWD-LSEGKHLFSLDGSCP-INSLGFFPCK---YWVGGATDKFFKIW 111
D LW+ L+E K+ + ++ + F P +V G DK K+W
Sbjct 139 DKTIKLWNTLAECKYTITNSTHTDWVSCVRFSPSGKEPIFVSGGWDKLIKVW 190
> bbo:BBOV_II003050 18.m06254; receptor for activated C kinase,
RACK protein; K14753 guanine nucleotide-binding protein subunit
beta-2-like 1 protein
Length=322
Score = 169 bits (429), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 82/164 (50%), Positives = 101/164 (61%), Gaps = 0/164 (0%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
DWVSCVRF P P E + GWDK+VKVW L C L NL G ++ ++SPDGSLCA
Sbjct 153 DWVSCVRFSPNPHEHVFVSGGWDKIVKVWDLANCNLKFNLSGHEGIVSCTSISPDGSLCA 212
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG+ LWD+ EG L L+ PIN+L F PC YW+ ATD+ KIW E K VL
Sbjct 213 SGGKDGIARLWDMKEGNSLHLLEAGSPINALCFSPCNYWLCVATDRAIKIWNLENKNVLA 272
Query 121 DISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
+I+ D P K G+PW W+ +G LF GS G I VY+V +
Sbjct 273 EITSDKPKKIGLPWCVSLCWNADGSTLFAGSTDGTIYVYQVNKN 316
Score = 42.0 bits (97), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 42/94 (44%), Gaps = 4/94 (4%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD-LSEGKHLF 80
WD +++W L CK G T +Y+V SPD +D LW+ LSE K
Sbjct 88 WDNTLRLWDLVKCKTVHVYNGHTSDVYSVDFSPDNRQIISASRDKTIKLWNTLSECKRTV 147
Query 81 SLDGSCPINSLGFFPCKY---WVGGATDKFFKIW 111
+ ++ + F P + +V G DK K+W
Sbjct 148 QNAHNDWVSCVRFSPNPHEHVFVSGGWDKIVKVW 181
> dre:30722 gnb2l1, rack1, wu:fb80d08, wu:fk65d12; guanine nucleotide
binding protein (G protein), beta polypeptide 2-like
1; K14753 guanine nucleotide-binding protein subunit beta-2-like
1 protein
Length=317
Score = 163 bits (412), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 84/167 (50%), Positives = 104/167 (62%), Gaps = 3/167 (1%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P+I CGWDK+VKVW+L CKL N +G T L TVTVSPDGSLCA
Sbjct 149 EWVSCVRFSPNSSNPIIVSCGWDKMVKVWNLANCKLKTNHIGHTGYLNTVTVSPDGSLCA 208
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG +LWDL+EGKHL++LDG IN+L F P +YW+ AT KIW E K ++D
Sbjct 209 SGGKDGQAMLWDLNEGKHLYTLDGGDTINALCFSPNRYWLCAATGPSIKIWDLEGKIIVD 268
Query 121 DISPD---HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ D K P WS +G+ LF G I V++V G
Sbjct 269 ELRQDIITTNSKAEPPQCTSLAWSADGQTLFAGYTDNLIRVWQVTIG 315
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 51/118 (43%), Gaps = 10/118 (8%)
Query 2 WVSCVRFFP-FPKEPLIFLCGWDKLVKVWSLTTCK----LPAN-LVGQTFVLYTVTVSPD 55
WV+ + P FP +I DK + +W LT + +P L G + + V +S D
Sbjct 17 WVTQIATTPQFP--DMILSASRDKTIIMWKLTRDETNYGIPQRALRGHSHFVSDVVISSD 74
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
G G DG LWDL+ G G + S+ F + V G+ DK K+W
Sbjct 75 GQFALSGSWDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSADNRQIVSGSRDKTIKLW 132
Score = 33.1 bits (74), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
WD +++W LTT VG T + +V S D G +D LW+
Sbjct 83 WDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSADNRQIVSGSRDKTIKLWN 133
> xla:495666 gnb2l1, gnb2-rs1, h12.3, hlc-7, pig21, rack1; guanine
nucleotide binding protein (G protein), beta polypeptide
2-like 1; K14753 guanine nucleotide-binding protein subunit
beta-2-like 1 protein
Length=317
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 83/167 (49%), Positives = 104/167 (62%), Gaps = 3/167 (1%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P+I CGWDKLVKVW+L CKL N +G T L TVTVSPDGSLCA
Sbjct 149 EWVSCVRFSPNSSNPIIVSCGWDKLVKVWNLANCKLKTNHIGHTGYLNTVTVSPDGSLCA 208
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG +LWDL+EGKHL++LDG IN+L F P +YW+ AT KIW E K ++D
Sbjct 209 SGGKDGQAMLWDLNEGKHLYTLDGGDIINALCFSPNRYWLCAATGPSIKIWDLEGKIIVD 268
Query 121 DISPD---HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ + K P WS +G+ LF G + V++V G
Sbjct 269 ELKQEVISTSSKAEPPQCTSLAWSADGQTLFAGYTDNLVRVWQVTIG 315
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 51/118 (43%), Gaps = 10/118 (8%)
Query 2 WVSCVRFFP-FPKEPLIFLCGWDKLVKVWSLTTCK----LPAN-LVGQTFVLYTVTVSPD 55
WV+ + P FP +I DK + +W LT + +P L G + + V +S D
Sbjct 17 WVTQIATTPQFP--DMILSASRDKTIIMWKLTRDETNYGIPQRALRGHSHFVSDVVISSD 74
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
G G DG LWDL+ G G + S+ F + V G+ DK K+W
Sbjct 75 GQFALSGSWDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSSDNRQIVSGSRDKTIKLW 132
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
WD +++W LTT VG T + +V S D G +D LW+
Sbjct 83 WDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSSDNRQIVSGSRDKTIKLWN 133
> mmu:14694 Gnb2l1, AL033335, GB-like, Gnb2-rs1, Rack1, p205;
guanine nucleotide binding protein (G protein), beta polypeptide
2 like 1; K14753 guanine nucleotide-binding protein subunit
beta-2-like 1 protein
Length=317
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 83/167 (49%), Positives = 104/167 (62%), Gaps = 3/167 (1%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P+I CGWDKLVKVW+L CKL N +G T L TVTVSPDGSLCA
Sbjct 149 EWVSCVRFSPNSSNPIIVSCGWDKLVKVWNLANCKLKTNHIGHTGYLNTVTVSPDGSLCA 208
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG +LWDL+EGKHL++LDG IN+L F P +YW+ AT KIW E K ++D
Sbjct 209 SGGKDGQAMLWDLNEGKHLYTLDGGDIINALCFSPNRYWLCAATGPSIKIWDLEGKIIVD 268
Query 121 DISPD---HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ + K P WS +G+ LF G + V++V G
Sbjct 269 ELKQEVISTSSKAEPPQCTSLAWSADGQTLFAGYTDNLVRVWQVTIG 315
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 51/118 (43%), Gaps = 10/118 (8%)
Query 2 WVSCVRFFP-FPKEPLIFLCGWDKLVKVWSLTTCK----LPAN-LVGQTFVLYTVTVSPD 55
WV+ + P FP +I DK + +W LT + +P L G + + V +S D
Sbjct 17 WVTQIATTPQFP--DMILSASRDKTIIMWKLTRDETNYGIPQRALRGHSHFVSDVVISSD 74
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
G G DG LWDL+ G G + S+ F + V G+ DK K+W
Sbjct 75 GQFALSGSWDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSSDNRQIVSGSRDKTIKLW 132
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
WD +++W LTT VG T + +V S D G +D LW+
Sbjct 83 WDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSSDNRQIVSGSRDKTIKLWN 133
> hsa:10399 GNB2L1, Gnb2-rs1, H12.3, HLC-7, PIG21, RACK1; guanine
nucleotide binding protein (G protein), beta polypeptide
2-like 1; K14753 guanine nucleotide-binding protein subunit
beta-2-like 1 protein
Length=317
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 83/167 (49%), Positives = 104/167 (62%), Gaps = 3/167 (1%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P+I CGWDKLVKVW+L CKL N +G T L TVTVSPDGSLCA
Sbjct 149 EWVSCVRFSPNSSNPIIVSCGWDKLVKVWNLANCKLKTNHIGHTGYLNTVTVSPDGSLCA 208
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG +LWDL+EGKHL++LDG IN+L F P +YW+ AT KIW E K ++D
Sbjct 209 SGGKDGQAMLWDLNEGKHLYTLDGGDIINALCFSPNRYWLCAATGPSIKIWDLEGKIIVD 268
Query 121 DISPD---HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ + K P WS +G+ LF G + V++V G
Sbjct 269 ELKQEVISTSSKAEPPQCTSLAWSADGQTLFAGYTDNLVRVWQVTIG 315
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 51/118 (43%), Gaps = 10/118 (8%)
Query 2 WVSCVRFFP-FPKEPLIFLCGWDKLVKVWSLTTCK----LPAN-LVGQTFVLYTVTVSPD 55
WV+ + P FP +I DK + +W LT + +P L G + + V +S D
Sbjct 17 WVTQIATTPQFP--DMILSASRDKTIIMWKLTRDETNYGIPQRALRGHSHFVSDVVISSD 74
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
G G DG LWDL+ G G + S+ F + V G+ DK K+W
Sbjct 75 GQFALSGSWDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSSDNRQIVSGSRDKTIKLW 132
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
WD +++W LTT VG T + +V S D G +D LW+
Sbjct 83 WDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSSDNRQIVSGSRDKTIKLWN 133
> xla:446289 MGC130692; hypothetical protein LOC446289; K00567
methylated-DNA-[protein]-cysteine S-methyltransferase [EC:2.1.1.63]
Length=317
Score = 158 bits (399), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 81/167 (48%), Positives = 103/167 (61%), Gaps = 3/167 (1%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P+I CGWDK+VKVW+L CKL N +G + L TVTVSPDGSLCA
Sbjct 149 EWVSCVRFSPNSSNPIIVSCGWDKMVKVWNLANCKLKTNHIGHSGYLNTVTVSPDGSLCA 208
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG +LWDL+EGKHL++LD IN+L F P +YW+ AT KIW E K ++D
Sbjct 209 SGGKDGQAMLWDLNEGKHLYTLDSGDVINALCFSPNRYWLCAATGPSIKIWDLEGKIIVD 268
Query 121 DISPD---HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ + K P WS +G+ LF G I V++V G
Sbjct 269 ELKQEVISSSSKAEPPQCTSLAWSADGQTLFAGYTDNLIRVWQVTIG 315
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 49/118 (41%), Gaps = 10/118 (8%)
Query 2 WVSCVRFFP-FPKEPLIFLCGWDKLVKVWSLTTCKL-----PANLVGQTFVLYTVTVSPD 55
WV+ + P FP +I DK V +W LT + L G + + V +S D
Sbjct 17 WVTQIATTPQFP--DMILSSSRDKSVIMWKLTRDETNYGVPQRALRGHSHFVSDVVISSD 74
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
G G DG LWDL+ G G + S+ F + V G+ DK K+W
Sbjct 75 GQFALSGSWDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSADNRQIVSGSRDKTIKLW 132
Score = 33.5 bits (75), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
WD +++W LTT VG T + +V S D G +D LW+
Sbjct 83 WDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSADNRQIVSGSRDKTIKLWN 133
> xla:379825 gnb2l1, MGC53289; guanine nucleotide binding protein,
beta 2, related sequence 1; K14753 guanine nucleotide-binding
protein subunit beta-2-like 1 protein
Length=317
Score = 158 bits (399), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 81/167 (48%), Positives = 103/167 (61%), Gaps = 3/167 (1%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P+I CGWDK+VKVW+L CKL N +G + L TVTVSPDGSLCA
Sbjct 149 EWVSCVRFSPNSSNPIIVSCGWDKMVKVWNLANCKLKTNHIGHSGYLNTVTVSPDGSLCA 208
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG +LWDL+EGKHL++LD IN+L F P +YW+ AT KIW E K ++D
Sbjct 209 SGGKDGQAMLWDLNEGKHLYTLDSGDVINALCFSPNRYWLCAATGPSIKIWDLEGKIIVD 268
Query 121 DISPD---HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
++ + K P WS +G+ LF G I V++V G
Sbjct 269 ELKQEVISSSSKAEPPQCTSLAWSADGQTLFAGYTDNLIRVWQVTIG 315
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 49/118 (41%), Gaps = 10/118 (8%)
Query 2 WVSCVRFFP-FPKEPLIFLCGWDKLVKVWSLTTCKL-----PANLVGQTFVLYTVTVSPD 55
WV+ + P FP +I DK V +W LT + L G + + V +S D
Sbjct 17 WVTQIATTPQFP--DMILSSSRDKSVIMWKLTRDETNYGVPQRALRGHSHFVSDVVISSD 74
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
G G DG LWDL+ G G + S+ F + V G+ DK K+W
Sbjct 75 GQFALSGSWDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSADNRQIVSGSRDKTIKLW 132
Score = 33.5 bits (75), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
WD +++W LTT VG T + +V S D G +D LW+
Sbjct 83 WDGTLRLWDLTTGTTTRRFVGHTKDVLSVAFSADNRQIVSGSRDKTIKLWN 133
> cel:K04D7.1 rack-1; RACK1 (mammalian Receptor of Activated C
Kinase) homolog family member (rack-1); K14753 guanine nucleotide-binding
protein subunit beta-2-like 1 protein
Length=325
Score = 154 bits (390), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 77/166 (46%), Positives = 104/166 (62%), Gaps = 4/166 (2%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
DWVS VRF P ++P+I GWDK+VKVW+L C+L N +G T + TVTVSPDGSLCA
Sbjct 155 DWVSTVRFSPSNRDPVIVSAGWDKVVKVWNLGNCRLKTNHIGHTGYVNTVTVSPDGSLCA 214
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDG +LWDL+EGKHL++L G+ IN++ F P +YW+ A KIW E KK ++
Sbjct 215 SGGKDGQAMLWDLNEGKHLYTLPGNDVINAMSFSPNRYWLCAAVGSSIKIWDLEDKKEIE 274
Query 121 DISPD----HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVA 162
++ P+ + P WS +G+ LF G I VY+V+
Sbjct 275 ELKPEIASSGSSRGSSPQCISLAWSQDGQTLFAGYTDNIIRVYQVS 320
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 39/166 (23%), Positives = 62/166 (37%), Gaps = 22/166 (13%)
Query 2 WVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTC---------KLPANLVGQTFVLYTVTV 52
WV+ + + + + DK + VW + + + +L G + V +
Sbjct 18 WVTQIATYTRNDKTTVLSSSRDKTILVWDVDSVAPVDEGPIGRPVRSLTGHNHFVSDVVI 77
Query 53 SPDGSLCAFGGKDGVTILWDLSEGKHLFS-LDGSCPINSLGFFP-CKYWVGGATDKFFKI 110
S DG G D LWDL++G + + + S+ F + V G+ DK K+
Sbjct 78 SSDGQFALSGSWDKTLRLWDLNQGVSTRQFISHTKDVLSVAFSADNRQIVSGSRDKSIKL 137
Query 111 W---APEKKKVLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAG 153
W A K + DD D W +S R I SAG
Sbjct 138 WNTLAQCKYTITDDCHTD--------WVSTVRFSPSNRDPVIVSAG 175
> ath:AT1G18080 ATARCA; ATARCA; nucleotide binding; K14753 guanine
nucleotide-binding protein subunit beta-2-like 1 protein
Length=327
Score = 154 bits (389), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 79/176 (44%), Positives = 103/176 (58%), Gaps = 13/176 (7%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
DWVSCVRF P +P I WDK VKVW+L+ CKL + L G T + TV VSPDGSLCA
Sbjct 151 DWVSCVRFSPNTLQPTIVSASWDKTVKVWNLSNCKLRSTLAGHTGYVSTVAVSPDGSLCA 210
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDGV +LWDL+EGK L+SL+ + I++L F P +YW+ AT+ KIW E K +++
Sbjct 211 SGGKDGVVLLWDLAEGKKLYSLEANSVIHALCFSPNRYWLCAATEHGIKIWDLESKSIVE 270
Query 121 DISPD-------------HPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAE 163
D+ D KR + + NWS +G LF G G I V+ +
Sbjct 271 DLKVDLKAEAEKADNSGPAATKRKVIYCTSLNWSADGSTLFSGYTDGVIRVWGIGR 326
Score = 35.8 bits (81), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 28/109 (25%), Positives = 41/109 (37%), Gaps = 7/109 (6%)
Query 10 PFPKEPLIFLCGWDKLVKVWSLTTCK-----LPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
P +I DK + +W LT L G + + V +S DG G
Sbjct 24 PIDNADIIVSASRDKSIILWKLTKDDKAYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSW 83
Query 65 DGVTILWDLSEGKHLFSLDGSCP-INSLGF-FPCKYWVGGATDKFFKIW 111
DG LWDL+ G G + S+ F + V + D+ K+W
Sbjct 84 DGELRLWDLAAGVSTRRFVGHTKDVLSVAFSLDNRQIVSASRDRTIKLW 132
> ath:AT3G18130 RACK1C_AT (RECEPTOR FOR ACTIVATED C KINASE 1 C);
nucleotide binding; K14753 guanine nucleotide-binding protein
subunit beta-2-like 1 protein
Length=326
Score = 146 bits (369), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 77/176 (43%), Positives = 101/176 (57%), Gaps = 13/176 (7%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P I WDK VKVW+L CKL +LVG + L TV VSPDGSLCA
Sbjct 150 EWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNSLVGHSGYLNTVAVSPDGSLCA 209
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDGV +LWDL+EGK L+SL+ I+SL F P +YW+ AT+ +IW E K V++
Sbjct 210 SGGKDGVILLWDLAEGKKLYSLEAGSIIHSLCFSPNRYWLCAATENSIRIWDLESKSVVE 269
Query 121 DISPDHPV-------------KRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAE 163
D+ D ++ + + NWS +G LF G G + V+ +
Sbjct 270 DLKVDLKSEAEKNEGGVGTGNQKKVIYCTSLNWSADGSTLFSGYTDGVVRVWGIGR 325
Score = 36.6 bits (83), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 28/109 (25%), Positives = 42/109 (38%), Gaps = 7/109 (6%)
Query 10 PFPKEPLIFLCGWDKLVKVWSLTTCK-----LPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
P +I DK + +W LT L G + + V +S DG G
Sbjct 24 PIDNSDIIVTASRDKSIILWKLTKDDKSYGVAQRRLTGHSHFVEDVVLSSDGQFALSGSW 83
Query 65 DGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
DG LWDL+ G+ G + S+ F + V + D+ K+W
Sbjct 84 DGELRLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLW 132
Score = 33.1 bits (74), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 60/149 (40%), Gaps = 12/149 (8%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD-LSEGKHLF 80
WD +++W L T + VG T + +V S D +D LW+ L E K+
Sbjct 83 WDGELRLWDLATGETTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLWNTLGECKYTI 142
Query 81 SL-DGSCP-INSLGFFPCKY---WVGGATDKFFKIWAPEKKKVLDDISPDHPVKRGIPWG 135
S DG ++ + F P V + DK K+W + K+ + + +
Sbjct 143 SEGDGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNSLVGHSGYLNTVAVS 202
Query 136 PFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
P +G G G I ++++AEG
Sbjct 203 P------DGSLCASGGKDGVILLWDLAEG 225
> ath:AT1G48630 RACK1B_AT (RECEPTOR FOR ACTIVATED C KINASE 1 B);
nucleotide binding; K14753 guanine nucleotide-binding protein
subunit beta-2-like 1 protein
Length=326
Score = 146 bits (368), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 78/176 (44%), Positives = 98/176 (55%), Gaps = 13/176 (7%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WVSCVRF P P I WDK VKVW+L CKL L G + L TV VSPDGSLCA
Sbjct 150 EWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNTLAGHSGYLNTVAVSPDGSLCA 209
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD 120
GGKDGV +LWDL+EGK L+SL+ I+SL F P +YW+ AT+ +IW E K V++
Sbjct 210 SGGKDGVILLWDLAEGKKLYSLEAGSIIHSLCFSPNRYWLCAATENSIRIWDLESKSVVE 269
Query 121 DISPDHPV-------------KRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAE 163
D+ D K + + NWS +G LF G G I V+ +
Sbjct 270 DLKVDLKAEAEKTDGSTGIGNKTKVIYCTSLNWSADGNTLFSGYTDGVIRVWGIGR 325
Score = 35.4 bits (80), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 42/109 (38%), Gaps = 7/109 (6%)
Query 10 PFPKEPLIFLCGWDKLVKVWSLTTCK-----LPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
P +I DK + +W LT + G + + V +S DG G
Sbjct 24 PVDNSDVIVTSSRDKSIILWKLTKEDKSYGVAQRRMTGHSHFVQDVVLSSDGQFALSGSW 83
Query 65 DGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
DG LWDL+ G+ G + S+ F + V + D+ K+W
Sbjct 84 DGELRLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLW 132
Score = 34.3 bits (77), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 61/149 (40%), Gaps = 12/149 (8%)
Query 22 WDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD-LSEGKHLF 80
WD +++W L T + VG T + +V S D +D LW+ L E K+
Sbjct 83 WDGELRLWDLATGESTRRFVGHTKDVLSVAFSTDNRQIVSASRDRTIKLWNTLGECKYTI 142
Query 81 S-LDGSCP-INSLGFFPCKY---WVGGATDKFFKIWAPEKKKVLDDISPDHPVKRGIPWG 135
S DG ++ + F P V + DK K+W + K+ + ++ +
Sbjct 143 SEADGHKEWVSCVRFSPNTLVPTIVSASWDKTVKVWNLQNCKLRNTLAGHSGYLNTVAVS 202
Query 136 PFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
P +G G G I ++++AEG
Sbjct 203 P------DGSLCASGGKDGVILLWDLAEG 225
> sce:YMR116C ASC1, CPC2; Asc1p; K14753 guanine nucleotide-binding
protein subunit beta-2-like 1 protein
Length=319
Score = 108 bits (271), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 58/167 (34%), Positives = 89/167 (53%), Gaps = 6/167 (3%)
Query 1 DWVSCVRFFPFPKEP----LIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDG 56
DWVS VR P K I G DK+VK W+L ++ A+ +G + T+T SPDG
Sbjct 149 DWVSQVRVVPNEKADDDSVTIISAGNDKMVKAWNLNQFQIEADFIGHNSNINTLTASPDG 208
Query 57 SLCAFGGKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKK 116
+L A GKDG +LW+L+ K +++L + SL F P +YW+ AT K+++ + +
Sbjct 209 TLIASAGKDGEIMLWNLAAKKAMYTLSAQDEVFSLAFSPNRYWLAAATATGIKVFSLDPQ 268
Query 117 KVLDDISPDHP--VKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEV 161
++DD+ P+ K P WS +G+ LF G I V++V
Sbjct 269 YLVDDLRPEFAGYSKAAEPHAVSLAWSADGQTLFAGYTDNVIRVWQV 315
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 22/110 (20%), Positives = 45/110 (40%), Gaps = 9/110 (8%)
Query 20 CGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHL 79
WDK +++W + T + VG + +V + S+ G +D +W + +G+ L
Sbjct 83 ASWDKTLRLWDVATGETYQRFVGHKSDVMSVDIDKKASMIISGSRDKTIKVWTI-KGQCL 141
Query 80 FSLDGSCP-INSLGFFPCK-------YWVGGATDKFFKIWAPEKKKVLDD 121
+L G ++ + P + + DK K W + ++ D
Sbjct 142 ATLLGHNDWVSQVRVVPNEKADDDSVTIISAGNDKMVKAWNLNQFQIEAD 191
> ath:AT1G61210 WD-40 repeat family protein / katanin p80 subunit,
putative
Length=1180
Score = 60.1 bits (144), Expect = 3e-09, Method: Composition-based stats.
Identities = 46/152 (30%), Positives = 67/152 (44%), Gaps = 16/152 (10%)
Query 4 SCVRFFPFPKEPLIFLCGW--DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAF 61
S V F PF + FL D +K+W + G + + T+ +PDG
Sbjct 104 SAVEFHPFGE----FLASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDGRWVVS 159
Query 62 GGKDGVTILWDLSEGK--HLFSLDGSCPINSLGFFPCKYWVG-GATDKFFKIWAPEKKKV 118
GG D V +WDL+ GK H F PI SL F P ++ + G+ D+ K W E ++
Sbjct 160 GGLDNVVKVWDLTAGKLLHEFKFHEG-PIRSLDFHPLEFLLATGSADRTVKFWDLETFEL 218
Query 119 LDDISPDHPVKRGIPWGPFFNWSYEGRPLFIG 150
+ P+ R I + P +GR LF G
Sbjct 219 IGSTRPEATGVRSIKFHP------DGRTLFCG 244
Score = 38.5 bits (88), Expect = 0.011, Method: Composition-based stats.
Identities = 30/109 (27%), Positives = 49/109 (44%), Gaps = 7/109 (6%)
Query 13 KEPLIFLCGWDKL-VKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILW 71
K +F+ G D V +W++ +L G T + +V L G GV LW
Sbjct 26 KTSRLFITGGDDYKVNLWAIGKPTSLMSLCGHTSAVDSVAFDSAEVLVLAGASSGVIKLW 85
Query 72 DLSEGKHLFSLDG---SCPINSLGFFPC-KYWVGGATDKFFKIWAPEKK 116
D+ E K + + G +C +++ F P ++ G++D KIW KK
Sbjct 86 DVEEAKMVRAFTGHRSNC--SAVEFHPFGEFLASGSSDANLKIWDIRKK 132
> ath:AT1G11160 nucleotide binding
Length=1021
Score = 58.9 bits (141), Expect = 8e-09, Method: Composition-based stats.
Identities = 48/157 (30%), Positives = 67/157 (42%), Gaps = 18/157 (11%)
Query 4 SCVRFFPFPKEPLIFLCGW--DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAF 61
S V F PF + FL D ++VW G T + T+ SPDG
Sbjct 104 SAVEFHPFGE----FLASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDGRWVVS 159
Query 62 GGKDGVTILWDLSEGKHLFSLDGSC---PINSLGFFPCKYWVG-GATDKFFKIWAPEKKK 117
GG D V +WDL+ GK L C PI SL F P ++ + G+ D+ K W E +
Sbjct 160 GGLDNVVKVWDLTAGKLLHEF--KCHEGPIRSLDFHPLEFLLATGSADRTVKFWDLETFE 217
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGG 154
++ P+ R I + P +G+ LF G G
Sbjct 218 LIGTTRPEATGVRAIAFHP------DGQTLFCGLDDG 248
Score = 45.8 bits (107), Expect = 6e-05, Method: Composition-based stats.
Identities = 40/153 (26%), Positives = 67/153 (43%), Gaps = 12/153 (7%)
Query 16 LIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSE 75
L+ G D V +WS+ P +L G T + +V + + L G GV LWDL E
Sbjct 30 LLLTGGDDYKVNLWSIGKTTSPMSLCGHTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEE 89
Query 76 GKHLFSLDG---SCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKKVLDDISPDHPVKRG 131
K + + G +C +++ F P ++ G++D ++W KK + RG
Sbjct 90 SKMVRAFTGHRSNC--SAVEFHPFGEFLASGSSDTNLRVWDTRKKGCIQTYKGH---TRG 144
Query 132 IPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
I F S +GR + G + V+++ G
Sbjct 145 ISTIEF---SPDGRWVVSGGLDNVVKVWDLTAG 174
> sce:YBR198C TAF5, TAF90; Taf5p; K03130 transcription initiation
factor TFIID subunit 5
Length=798
Score = 54.7 bits (130), Expect = 1e-07, Method: Composition-based stats.
Identities = 35/128 (27%), Positives = 55/128 (42%), Gaps = 5/128 (3%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
V CV F P +F DK ++W ++T +G T + ++ V PDG + G
Sbjct 612 VDCVSFHP--NGCYVFTGSSDKTCRMWDVSTGDSVRLFLGHTAPVISIAVCPDGRWLSTG 669
Query 63 GKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFP-CKYWVGGATDKFFKIWAPEKKKVL 119
+DG+ +WD+ GK L + G I SL + + G D ++W +K
Sbjct 670 SEDGIINVWDIGTGKRLKQMRGHGKNAIYSLSYSKEGNVLISGGADHTVRVWDLKKATTE 729
Query 120 DDISPDHP 127
PD P
Sbjct 730 PSAEPDEP 737
Score = 39.3 bits (90), Expect = 0.006, Method: Composition-based stats.
Identities = 28/112 (25%), Positives = 46/112 (41%), Gaps = 25/112 (22%)
Query 23 DKLVKVWSLT--------------------TCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
D +K+WSL TCK LVG + +Y+ + SPD G
Sbjct 487 DSYIKIWSLDGSSLNNPNIALNNNDKDEDPTCK---TLVGHSGTVYSTSFSPDNKYLLSG 543
Query 63 GKDGVTILWDLSEGKHLFSLDG-SCPINSLGFFPCKYWVGGAT-DKFFKIWA 112
+D LW + L S G + P+ + F P ++ A+ D+ ++W+
Sbjct 544 SEDKTVRLWSMDTHTALVSYKGHNHPVWDVSFSPLGHYFATASHDQTARLWS 595
> dre:386932 poc1b, TUWD12, fl36w17, wdr51b, wu:fl36w17, zgc:63538;
POC1 centriolar protein homolog B (Chlamydomonas)
Length=490
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 46/160 (28%), Positives = 68/160 (42%), Gaps = 12/160 (7%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
D +SC F P K+ C DK + +W+L VG T V+ V +P GSL A
Sbjct 19 DVISCADFNPNNKQLATGSC--DKSLMIWNLAPKARAFRFVGHTDVITGVNFAPSGSLVA 76
Query 61 FGGKDGVTILWDLS-EGKHLFSLDGSCPINSLGF-FPCKYWVGGATDKFFKIWAPEKKKV 118
+D LW S +G+ + + S+ F + V + DK K+W E+KK
Sbjct 77 SSSRDQTVRLWTPSIKGESTVFKAHTASVRSVNFSRDGQRLVTASDDKSVKVWGVERKKF 136
Query 119 LDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPV 158
L + + R W +S +GR I S G + V
Sbjct 137 L------YSLNRHTNWVRCARFSPDGR--LIASCGDDRTV 168
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 47/112 (41%), Gaps = 4/112 (3%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
+WV C RF P + LI CG D+ V++W ++ + V + G+ A
Sbjct 145 NWVRCARFSPDGR--LIASCGDDRTVRLWDTSSHQCTNIFTDYGGSATFVDFNSSGTCIA 202
Query 61 FGGKDGVTILWDLSEGKHLFSLD-GSCPINSLGFFPC-KYWVGGATDKFFKI 110
G D +WD+ K + + +N F P Y + G++D KI
Sbjct 203 SSGADNTIKIWDIRTNKLIQHYKVHNAGVNCFSFHPSGNYLISGSSDSTIKI 254
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 44/169 (26%), Positives = 64/169 (37%), Gaps = 17/169 (10%)
Query 3 VSCVRFFPFPKE-PLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAF 61
+ VR F ++ + DK VKVW + K +L T + SPDG L A
Sbjct 102 TASVRSVNFSRDGQRLVTASDDKSVKVWGVERKKFLYSLNRHTNWVRCARFSPDGRLIAS 161
Query 62 GGKDGVTILWDLSEGK--HLFSLDGSCPI----NSLGFFPCKYWVGGATDKFFKIWAPEK 115
G D LWD S + ++F+ G NS G C G D KIW
Sbjct 162 CGDDRTVRLWDTSSHQCTNIFTDYGGSATFVDFNSSG--TCIASSGA--DNTIKIWDIRT 217
Query 116 KKVLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
K++ + + P N+ L GS+ I + ++ EG
Sbjct 218 NKLIQHYKVHNAGVNCFSFHPSGNY------LISGSSDSTIKILDLLEG 260
Score = 33.9 bits (76), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 30/69 (43%), Gaps = 2/69 (2%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
V+C F P + D +K+ L +L L G + TVT S DG L A G
Sbjct 231 VNCFSFHPSGN--YLISGSSDSTIKILDLLEGRLIYTLHGHKGPVLTVTFSRDGDLFASG 288
Query 63 GKDGVTILW 71
G D ++W
Sbjct 289 GADSQVLMW 297
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 30/68 (44%), Gaps = 0/68 (0%)
Query 17 IFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEG 76
I G D +K+W + T KL + + + P G+ G D + DL EG
Sbjct 201 IASSGADNTIKIWDIRTNKLIQHYKVHNAGVNCFSFHPSGNYLISGSSDSTIKILDLLEG 260
Query 77 KHLFSLDG 84
+ +++L G
Sbjct 261 RLIYTLHG 268
> cpv:cgd7_3960 pleiotropic regulator 1 ; K12862 pleiotropic regulator
1
Length=427
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 53/113 (46%), Gaps = 5/113 (4%)
Query 3 VSCVRFFPF-PKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAF 61
++ VR F + P +F C DK +K W L ++ N + +Y + + P + A
Sbjct 173 IAAVRKVLFSERHPFLFSCSEDKTMKCWDLEQNRIVRNYARHSSGIYCLDIHPRLDIVAT 232
Query 62 GGKDGVTILWDL--SEGKHLFSLDGSCPINSLGFFPCK-YWVGGATDKFFKIW 111
G +DG +LWD+ E HLF + I+S+ + + G+ D+ + W
Sbjct 233 GSRDGSVVLWDIRTRESIHLFK-NHKAAISSILMQSIEPQLISGSYDRTIRTW 284
> xla:398057 paaf1, paaf, rpn14, wdr71; proteasomal ATPase-associated
factor 1; K11887 proteasomal ATPase-associated factor
1
Length=376
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/115 (31%), Positives = 53/115 (46%), Gaps = 7/115 (6%)
Query 5 CVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
C +FFP +E + G D LVKVWS+ A L G + + V DG G+
Sbjct 116 CCKFFPSGQE--VLSGGLDSLVKVWSVNDGSCLATLKGHRGSILDIAVVADGQNVISSGQ 173
Query 65 DGVTILWDLSEGKHLFSLDGS-CPINSLGFFPCKYWVGGATDKFFKIWAPEKKKV 118
DG LWD ++G + +D S PIN++ VG + + AP ++V
Sbjct 174 DGTARLWDSAQGSCISVVDDSYSPINAIAVGE----VGNSVNLGSSKEAPSDREV 224
Score = 32.3 bits (72), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 29/62 (46%), Gaps = 0/62 (0%)
Query 23 DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSL 82
D+ KVW ++ + L G T ++ P G GG D + +W +++G L +L
Sbjct 90 DQTFKVWETHNAEVKSVLEGHTMDVFCCKFFPSGQEVLSGGLDSLVKVWSVNDGSCLATL 149
Query 83 DG 84
G
Sbjct 150 KG 151
> cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=492
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/100 (34%), Positives = 49/100 (49%), Gaps = 3/100 (3%)
Query 5 CVRFFPFPKEPLIFLCGW-DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
V F + LIF G D +VK+W L + A G T + ++ S +G A G
Sbjct 340 AVHSIEFHPDGLIFGTGAADAVVKIWDLKNQTVAAAFPGHTAAVRSIAFSENGYYLATGS 399
Query 64 KDGVTILWDLSEGKHL--FSLDGSCPINSLGFFPCKYWVG 101
+DG LWDL + K+L F+ + PINSL F ++G
Sbjct 400 EDGEVKLWDLRKLKNLKTFANEEKQPINSLSFDMTGTFLG 439
Score = 37.4 bits (85), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 28/122 (22%), Positives = 49/122 (40%), Gaps = 9/122 (7%)
Query 42 GQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSLDG-SCPINSLGFFPCKYWV 100
G ++++ PDG + G D V +WDL + G + + S+ F Y++
Sbjct 336 GSQIAVHSIEFHPDGLIFGTGAADAVVKIWDLKNQTVAAAFPGHTAAVRSIAFSENGYYL 395
Query 101 G-GATDKFFKIWAPEKKKVLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVY 159
G+ D K+W K K L + + P + S++ F+G G + V
Sbjct 396 ATGSEDGEVKLWDLRKLKNLKTFANEEK-------QPINSLSFDMTGTFLGIGGQKVQVL 448
Query 160 EV 161
V
Sbjct 449 HV 450
> pfa:PFC0100c regulatory protein, putative; K12862 pleiotropic
regulator 1
Length=600
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 46/103 (44%), Gaps = 6/103 (5%)
Query 13 KEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
K P +F CG D VK W L K+ + G +Y +++ P L GG+D V +WD
Sbjct 339 KNPYLFSCGEDNRVKCWDLEYNKVIRDYHGHLSGVYCLSLHPSLDLLMSGGRDAVVRVWD 398
Query 73 LSEGKHLFSLDGSC----PINSLGFFPCKYWVGGATDKFFKIW 111
+ +F L G I S P V G+ DK ++W
Sbjct 399 IRTKSSVFVLSGHTGTVMSICSQSVEP--QVVSGSQDKMIRLW 439
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 39/156 (25%), Positives = 62/156 (39%), Gaps = 6/156 (3%)
Query 12 PKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILW 71
P L+ G D +V+VW + T L G T + ++ G +D + LW
Sbjct 380 PSLDLLMSGGRDAVVRVWDIRTKSSVFVLSGHTGTVMSICSQSVEPQVVSGSQDKMIRLW 439
Query 72 DLSEGKHLFSLD-GSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLDDISPDHPVKR 130
DL+ GK SL I SL P +Y K+W + +I+ + +
Sbjct 440 DLNNGKCRISLTHHKKSIRSLSIHPFEYSFCSCGTDNVKVWCGADAEFDRNITGFNSIIN 499
Query 131 G--IPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
I P+FN + L +GS G + Y+ + G
Sbjct 500 CSLIKQDPYFN---DSSILILGSNNGQLHFYDWSSG 532
Score = 38.1 bits (87), Expect = 0.012, Method: Composition-based stats.
Identities = 23/91 (25%), Positives = 36/91 (39%), Gaps = 2/91 (2%)
Query 23 DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSL 82
D+L+K+W L +CKL L G + + +S G+D WDL K +
Sbjct 307 DRLIKIWDLASCKLKLTLTGHINSIRDIKISKKNPYLFSCGEDNRVKCWDLEYNKVIRDY 366
Query 83 DGSCP-INSLGFFPC-KYWVGGATDKFFKIW 111
G + L P + G D ++W
Sbjct 367 HGHLSGVYCLSLHPSLDLLMSGGRDAVVRVW 397
> mmu:72137 Wdsub1, 1700048E19Rik, 2610014F08Rik; WD repeat, SAM
and U-box domain containing 1
Length=423
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 45/165 (27%), Positives = 70/165 (42%), Gaps = 15/165 (9%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTT-CKLPAN-LVGQTFVLYTVTVSPDGSL 58
D VSC F L+ C DK ++++SL+ +LP + L T+ ++ SP G +
Sbjct 13 DDVSCCAF----SAALLATCSLDKTIRLYSLSDFAELPYSPLKFHTYAVHCCCFSPSGHV 68
Query 59 CAFGGKDGVTILWDLSEGKHLFSLD--GSCPINSLGFFP-CKYWVGGATDKFFKIWAPEK 115
A DG T+LW G L L+ G P+ F P Y GA D +W +
Sbjct 69 LASCSTDGTTVLWSSHSGHTLTVLEQPGGSPVRVCCFSPDSAYLASGAADGSIALWNAQT 128
Query 116 KKVLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYE 160
K+ S + P +G GS+GG++ V++
Sbjct 129 YKLYRCGSVKDSSLVACAFSP------DGGLFVTGSSGGDLTVWD 167
> tgo:TGME49_018420 pleiotropic regulator 1, putative (EC:2.7.11.7);
K12862 pleiotropic regulator 1
Length=576
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/101 (28%), Positives = 48/101 (47%), Gaps = 2/101 (1%)
Query 13 KEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
+ P +F CG D VK W L K+ + G +YT+ + P + GG+D V +WD
Sbjct 317 RHPYMFTCGEDNRVKCWDLEQNKVVRDYHGHLSGVYTLALHPQLDILCSGGRDAVVRVWD 376
Query 73 LSEGKHLFSLDG-SCPINSLGFFPCK-YWVGGATDKFFKIW 111
+ ++ L G I SL + + + G+ DK ++W
Sbjct 377 MRTKHEIYVLSGHQGTIMSLQMQALEPHIISGSQDKMVRLW 417
Score = 33.5 bits (75), Expect = 0.29, Method: Composition-based stats.
Identities = 27/113 (23%), Positives = 45/113 (39%), Gaps = 6/113 (5%)
Query 2 WVSCVRFFPFPKEPLIFLCGW-DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
WV+C+ P + F G D+L+K+W L + L +L G + + +S
Sbjct 266 WVTCLAVDPTNE---WFATGSNDRLIKIWDLASGTLKLSLTGHVSAIRDIKISSRHPYMF 322
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFP-CKYWVGGATDKFFKIW 111
G+D WDL + K + G + +L P G D ++W
Sbjct 323 TCGEDNRVKCWDLEQNKVVRDYHGHLSGVYTLALHPQLDILCSGGRDAVVRVW 375
> cel:C14B1.4 tag-125; Temporarily Assigned Gene name family member
(tag-125); K14963 COMPASS component SWD3
Length=376
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 67/147 (45%), Gaps = 9/147 (6%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
++V C F P + L+ +D+ V++W + T L + + V+ + DGSL A
Sbjct 172 NYVFCCNFNP--QSSLVVSGSFDESVRIWDVKTGMCIKTLPAHSDPVSAVSFNRDGSLIA 229
Query 61 FGGKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKK 117
G DG+ +WD + G+ + +L D + P+ + F P KY + D K+W K K
Sbjct 230 SGSYDGLVRIWDTANGQCIKTLVDDENPPVAFVKFSPNGKYILASNLDSTLKLWDFSKGK 289
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEG 144
L + K I F N+S G
Sbjct 290 TLKQYTGHENSKYCI----FANFSVTG 312
Score = 33.9 bits (76), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 36/159 (22%), Positives = 58/159 (36%), Gaps = 26/159 (16%)
Query 5 CVRFFPFPKEP-----------LIFLCGWDKLVKVWSLTTCKLPANLVG-QTFVLYTVTV 52
C++ P +P LI +D LV++W + LV + + V
Sbjct 205 CIKTLPAHSDPVSAVSFNRDGSLIASGSYDGLVRIWDTANGQCIKTLVDDENPPVAFVKF 264
Query 53 SPDGSLCAFGGKDGVTILWDLSEGKHLFSLDGS-----CPINSLGFFPCKYWVGGATDKF 107
SP+G D LWD S+GK L G C + K+ + G+ D
Sbjct 265 SPNGKYILASNLDSTLKLWDFSKGKTLKQYTGHENSKYCIFANFSVTGGKWIISGSEDCK 324
Query 108 FKIWAPEKKKVLDDI---------SPDHPVKRGIPWGPF 137
IW + ++++ + S HPV+ I G
Sbjct 325 IYIWNLQTREIVQCLEGHTQPVLASDCHPVQNIIASGAL 363
> ath:AT5G13480 FY; FY; protein binding
Length=647
Score = 49.7 bits (117), Expect = 4e-06, Method: Composition-based stats.
Identities = 31/80 (38%), Positives = 39/80 (48%), Gaps = 1/80 (1%)
Query 6 VRFFPFPKEPLIFL-CGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
+R F K L F C D VKVW T C ++L G + + +V P SL GGK
Sbjct 210 IRDLSFCKTDLKFCSCSDDTTVKVWDFTKCVDESSLTGHGWDVKSVDWHPTKSLLVSGGK 269
Query 65 DGVTILWDLSEGKHLFSLDG 84
D + LWD G+ L SL G
Sbjct 270 DQLVKLWDTRSGRELCSLHG 289
Score = 42.4 bits (98), Expect = 7e-04, Method: Composition-based stats.
Identities = 29/103 (28%), Positives = 51/103 (49%), Gaps = 3/103 (2%)
Query 12 PKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILW 71
P + L+ G D+LVK+W + + +L G ++ +V + +G+ KD + L+
Sbjct 259 PTKSLLVSGGKDQLVKLWDTRSGRELCSLHGHKNIVLSVKWNQNGNWLLTASKDQIIKLY 318
Query 72 DLSEGKHLFSLDGSCP-INSLGFFPC--KYWVGGATDKFFKIW 111
D+ K L S G + SL + PC +Y+V G++D W
Sbjct 319 DIRTMKELQSFRGHTKDVTSLAWHPCHEEYFVSGSSDGSICHW 361
> mmu:69544 Wdr5b, 2310009C03Rik, AI606931; WD repeat domain 5B;
K14963 COMPASS component SWD3
Length=328
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 42/159 (26%), Positives = 72/159 (45%), Gaps = 9/159 (5%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
D+V C F P LI +D+ VK+W + T K L + + V + +GSL
Sbjct 124 DFVFCCDFNP--PSNLIVSGSFDESVKIWEVKTGKCLKTLSAHSDPISAVNFNCNGSLIV 181
Query 61 FGGKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKK 117
G DG+ +WD + G+ L +L +G+ P++ + F P KY + D K+W + +
Sbjct 182 SGSYDGLCRIWDAASGQCLRTLADEGNPPVSFVKFSPNGKYILTATLDNTLKLWDYSRGR 241
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNI 156
L + K + F ++S GR + + N+
Sbjct 242 CLKTYTGHKNEKYCL----FASFSVTGRKWVVSGSEDNM 276
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 34/145 (23%), Positives = 64/145 (44%), Gaps = 9/145 (6%)
Query 23 DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSL 82
DK +KVW + + K L G + ++ +P +L G D +W++ GK L +L
Sbjct 102 DKTLKVWDMRSGKCLKTLKGHSDFVFCCDFNPPSNLIVSGSFDESVKIWEVKTGKCLKTL 161
Query 83 DG-SCPINSLGFFPCK--YWVGGATDKFFKIWAPEKKKVLDDISPDHPVKRGIPWGPFFN 139
S PI+++ F C V G+ D +IW + L ++ + G P F
Sbjct 162 SAHSDPISAVN-FNCNGSLIVSGSYDGLCRIWDAASGQCLRTLADE-----GNPPVSFVK 215
Query 140 WSYEGRPLFIGSAGGNIPVYEVAEG 164
+S G+ + + + +++ + G
Sbjct 216 FSPNGKYILTATLDNTLKLWDYSRG 240
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 40/168 (23%), Positives = 61/168 (36%), Gaps = 18/168 (10%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
+S V+F P + D L+ +W L G + + V S D S
Sbjct 42 ISSVKFSP--NGEWLASSAADALIIIWGAYDGNCKKTLYGHSLEISDVAWSSDSSRLVSA 99
Query 63 GKDGVTILWDLSEGKHLFSLDGS------CPINSLGFFPCKYWVGGATDKFFKIWAPEKK 116
D +WD+ GK L +L G C N P V G+ D+ KIW +
Sbjct 100 SDDKTLKVWDMRSGKCLKTLKGHSDFVFCCDFNP----PSNLIVSGSFDESVKIWEVKTG 155
Query 117 KVLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
K L +S + N++ G + GS G +++ A G
Sbjct 156 KCLKTLSAHSDPISAV------NFNCNGSLIVSGSYDGLCRIWDAASG 197
Score = 33.9 bits (76), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 33/143 (23%), Positives = 51/143 (35%), Gaps = 23/143 (16%)
Query 16 LIFLCGWDKLVKVWSLTTCKLPANLVGQ-----TFVLYTVTVSPDGSLCAFGGKDGVTIL 70
LI +D L ++W + + L + +FV + SP+G D L
Sbjct 179 LIVSGSYDGLCRIWDAASGQCLRTLADEGNPPVSFVKF----SPNGKYILTATLDNTLKL 234
Query 71 WDLSEGKHLFSLDGS-----CPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLDDISPD 125
WD S G+ L + G C S K+ V G+ D IW + K+++ +
Sbjct 235 WDYSRGRCLKTYTGHKNEKYCLFASFSVTGRKWVVSGSEDNMVYIWNLQTKEIVQRLQGH 294
Query 126 ---------HPVKRGIPWGPFFN 139
HP K I N
Sbjct 295 TDVVISAACHPTKNIIASAALEN 317
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 36/86 (41%), Gaps = 2/86 (2%)
Query 36 LPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSLDG-SCPINSLGFF 94
L L G + + +V SP+G A D + I+W +G +L G S I+ + +
Sbjct 31 LRLTLAGHSAAISSVKFSPNGEWLASSAADALIIIWGAYDGNCKKTLYGHSLEISDVAWS 90
Query 95 P-CKYWVGGATDKFFKIWAPEKKKVL 119
V + DK K+W K L
Sbjct 91 SDSSRLVSASDDKTLKVWDMRSGKCL 116
> xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 41/159 (25%), Positives = 70/159 (44%), Gaps = 9/159 (5%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
++V C F P + LI +D+ V++W + T K L + + V + DGSL
Sbjct 130 NYVFCCNFNP--QSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSAVHFNRDGSLIV 187
Query 61 FGGKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKK 117
DG+ +WD + G+ L +L D + P++ + F P KY + D K+W K K
Sbjct 188 SSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGK 247
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNI 156
L + + + F N+S G + + N+
Sbjct 248 CL----KTYTCHKNEKYCIFANFSVTGGKWIVSGSEDNL 282
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 54/130 (41%), Gaps = 13/130 (10%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
VS V+F P + DKL+K+W K + G + V S D +L
Sbjct 48 VSSVKFSP--NGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLLVSA 105
Query 63 GKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKY------WVGGATDKFFKIWAPEKK 116
D +WD+S GK L +L G +S F C + V G+ D+ +IW +
Sbjct 106 SDDKTLKIWDVSSGKCLKTLKG----HSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTG 161
Query 117 KVLDDISPDH 126
K L + P H
Sbjct 162 KCLKTL-PAH 170
> xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat domain
5; K14963 COMPASS component SWD3
Length=334
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 41/159 (25%), Positives = 70/159 (44%), Gaps = 9/159 (5%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
++V C F P + LI +D+ V++W + T K L + + V + DGSL
Sbjct 130 NYVFCCNFNP--QSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSAVHFNRDGSLIV 187
Query 61 FGGKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKK 117
DG+ +WD + G+ L +L D + P++ + F P KY + D K+W K K
Sbjct 188 SSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGK 247
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNI 156
L + + + F N+S G + + N+
Sbjct 248 CL----KTYTCHKNEKYCIFANFSVTGGKWIVSGSEDNL 282
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 54/130 (41%), Gaps = 13/130 (10%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
VS V+F P + DKL+K+W K + G + V S D +L
Sbjct 48 VSSVKFSP--NGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLLVSA 105
Query 63 GKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKY------WVGGATDKFFKIWAPEKK 116
D +WD+S GK L +L G +S F C + V G+ D+ +IW +
Sbjct 106 SDDKTLKIWDISSGKCLKTLKG----HSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTG 161
Query 117 KVLDDISPDH 126
K L + P H
Sbjct 162 KCLKTL-PAH 170
> dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 70/159 (44%), Gaps = 9/159 (5%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
++V C F P + LI +D+ V++W + T K L + + V + DGSL
Sbjct 130 NYVFCCNFNP--QSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSAVHFNRDGSLIV 187
Query 61 FGGKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKK 117
DG+ +WD + G+ L +L D + P++ + F P KY + D K+W K K
Sbjct 188 SSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGK 247
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNI 156
L + K I F N+S G + + N+
Sbjct 248 CLKTYTGHKNEKYCI----FANFSVTGGKWIVSGSEDNM 282
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 55/130 (42%), Gaps = 13/130 (10%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
VS V+F P + + DKL+K+W K + G + V S D +L
Sbjct 48 VSSVKFSPSGE--WLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLLVSA 105
Query 63 GKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKY------WVGGATDKFFKIWAPEKK 116
D +WD+S GK L +L G +S F C + V G+ D+ +IW +
Sbjct 106 SDDKTLKIWDVSSGKCLKTLKG----HSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTG 161
Query 117 KVLDDISPDH 126
K L + P H
Sbjct 162 KCLKTL-PAH 170
> mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3;
WD repeat domain 5; K14963 COMPASS component SWD3
Length=334
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 70/159 (44%), Gaps = 9/159 (5%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
++V C F P + LI +D+ V++W + T K L + + V + DGSL
Sbjct 130 NYVFCCNFNP--QSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSAVHFNRDGSLIV 187
Query 61 FGGKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKK 117
DG+ +WD + G+ L +L D + P++ + F P KY + D K+W K K
Sbjct 188 SSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGK 247
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNI 156
L + K I F N+S G + + N+
Sbjct 248 CLKTYTGHKNEKYCI----FANFSVTGGKWIVSGSEDNL 282
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 54/130 (41%), Gaps = 13/130 (10%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
VS V+F P + DKL+K+W K + G + V S D +L
Sbjct 48 VSSVKFSP--NGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLLVSA 105
Query 63 GKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKY------WVGGATDKFFKIWAPEKK 116
D +WD+S GK L +L G +S F C + V G+ D+ +IW +
Sbjct 106 SDDKTLKIWDVSSGKCLKTLKG----HSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTG 161
Query 117 KVLDDISPDH 126
K L + P H
Sbjct 162 KCLKTL-PAH 170
> mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar protein
homolog B (Chlamydomonas)
Length=476
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 47/111 (42%), Gaps = 4/111 (3%)
Query 2 WVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAF 61
WV C +F P + LI C DK +K+W T + N V +P+G+ A
Sbjct 146 WVRCAKFSPDGR--LIVSCSEDKTIKIWDTTNKQCVNNFSDSVGFANFVDFNPNGTCIAS 203
Query 62 GGKDGVTILWDLSEGKHLFSLD-GSCPINSLGFFPC-KYWVGGATDKFFKI 110
G D +WD+ K L SC +N L F P V ++D K+
Sbjct 204 AGSDHAVKIWDIRMNKLLQHYQVHSCGVNCLSFHPLGNSLVTASSDGTVKM 254
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 39/162 (24%), Positives = 68/162 (41%), Gaps = 10/162 (6%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
++ F P K+ I WD + +WSL VG V+ ++ SP G+L A
Sbjct 21 ITSADFSPNCKQ--IATASWDTFLMLWSLKPHARAYRYVGHKDVVTSLQFSPQGNLLASA 78
Query 63 GKDGVTILWDL-SEGKHLFSLDGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKKVLD 120
+D LW L +GK + P+ S+ F + V + DK K+W+ +++ L
Sbjct 79 SRDRTVRLWVLDRKGKSSEFKAHTAPVRSVDFSADGQLLVTASEDKSIKVWSMFRQRFL- 137
Query 121 DISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVA 162
+ + R W +S +GR + S I +++
Sbjct 138 -----YSLYRHTHWVRCAKFSPDGRLIVSCSEDKTIKIWDTT 174
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 38/162 (23%), Positives = 62/162 (38%), Gaps = 9/162 (5%)
Query 6 VRFFPFPKE-PLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGK 64
VR F + L+ DK +KVWS+ + +L T + SPDG L +
Sbjct 105 VRSVDFSADGQLLVTASEDKSIKVWSMFRQRFLYSLYRHTHWVRCAKFSPDGRLIVSCSE 164
Query 65 DGVTILWDLSEGKHLFSLDGSCP-INSLGFFPCKYWVGGA-TDKFFKIWAPEKKKVLDDI 122
D +WD + + + + S N + F P + A +D KIW K+L
Sbjct 165 DKTIKIWDTTNKQCVNNFSDSVGFANFVDFNPNGTCIASAGSDHAVKIWDIRMNKLLQHY 224
Query 123 SPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
+ + P G L S+ G + + ++ EG
Sbjct 225 QVHSCGVNCLSFHPL------GNSLVTASSDGTVKMLDLIEG 260
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 35/133 (26%), Positives = 53/133 (39%), Gaps = 10/133 (7%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
V+C+ F P + D VK+ L +L L G T ++TV+ S DG L G
Sbjct 231 VNCLSFHPLGNS--LVTASSDGTVKMLDLIEGRLIYTLQGHTGPVFTVSFSKDGELLTSG 288
Query 63 GKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG-------GATDKFFKIWAPEK 115
G D ++W + HL D + L F + + D+ I K
Sbjct 289 GADAQVLIW-RTNFIHLHCKDPKRNLKRLHFEASPHLLDIYPRSPHSHEDRKETIEINPK 347
Query 116 KKVLDDISPDHPV 128
++V+D S PV
Sbjct 348 REVMDLQSSSPPV 360
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 37/160 (23%), Positives = 66/160 (41%), Gaps = 12/160 (7%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
D V+ ++F P + L+ D+ V++W L + T + +V S DG L
Sbjct 61 DVVTSLQFSP--QGNLLASASRDRTVRLWVLDRKGKSSEFKAHTAPVRSVDFSADGQLLV 118
Query 61 FGGKDGVTILWDLSEGKHLFSLDGSCP-INSLGFFPC-KYWVGGATDKFFKIWAPEKKKV 118
+D +W + + L+SL + F P + V + DK KIW K+
Sbjct 119 TASEDKSIKVWSMFRQRFLYSLYRHTHWVRCAKFSPDGRLIVSCSEDKTIKIWDTTNKQC 178
Query 119 LDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPV 158
+++ S + + F +++ G I SAG + V
Sbjct 179 VNNFSD------SVGFANFVDFNPNG--TCIASAGSDHAV 210
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 46/114 (40%), Gaps = 3/114 (2%)
Query 1 DWVSCVRFFPF-PKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLC 59
D V F F P I G D VK+W + KL + + + ++ P G+
Sbjct 184 DSVGFANFVDFNPNGTCIASAGSDHAVKIWDIRMNKLLQHYQVHSCGVNCLSFHPLGNSL 243
Query 60 AFGGKDGVTILWDLSEGKHLFSLDGSC-PINSLGFFPC-KYWVGGATDKFFKIW 111
DG + DL EG+ +++L G P+ ++ F + G D IW
Sbjct 244 VTASSDGTVKMLDLIEGRLIYTLQGHTGPVFTVSFSKDGELLTSGGADAQVLIW 297
> hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPASS
component SWD3
Length=334
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 70/159 (44%), Gaps = 9/159 (5%)
Query 1 DWVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCA 60
++V C F P + LI +D+ V++W + T K L + + V + DGSL
Sbjct 130 NYVFCCNFNP--QSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSDPVSAVHFNRDGSLIV 187
Query 61 FGGKDGVTILWDLSEGKHLFSL--DGSCPINSLGFFPC-KYWVGGATDKFFKIWAPEKKK 117
DG+ +WD + G+ L +L D + P++ + F P KY + D K+W K K
Sbjct 188 SSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGK 247
Query 118 VLDDISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNI 156
L + K I F N+S G + + N+
Sbjct 248 CLKTYTGHKNEKYCI----FANFSVTGGKWIVSGSEDNL 282
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 54/130 (41%), Gaps = 13/130 (10%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
VS V+F P + DKL+K+W K + G + V S D +L
Sbjct 48 VSSVKFSP--NGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSNLLVSA 105
Query 63 GKDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKY------WVGGATDKFFKIWAPEKK 116
D +WD+S GK L +L G +S F C + V G+ D+ +IW +
Sbjct 106 SDDKTLKIWDVSSGKCLKTLKG----HSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTG 161
Query 117 KVLDDISPDH 126
K L + P H
Sbjct 162 KCLKTL-PAH 170
> sce:YMR146C TIF34; Tif34p; K03246 translation initiation factor
3 subunit I
Length=347
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 37/160 (23%), Positives = 62/160 (38%), Gaps = 13/160 (8%)
Query 16 LIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSE 75
L+F C D VW + L G T ++++ V C G D LWD+S
Sbjct 24 LLFSCSKDSSASVWYSLNGERLGTLDGHTGTIWSIDVDCFTKYCVTGSADYSIKLWDVSN 83
Query 76 GKHLFSLDGSCPINSLGFFPCKYWVGGATDKFFK------IWAPEKKKVLDDIS-----P 124
G+ + + P+ + F PC + D K I+ E+ +++ P
Sbjct 84 GQCVATWKSPVPVKRVEFSPCGNYFLAILDNVMKNPGSINIYEIERDSATHELTKVSEEP 143
Query 125 DHPV--KRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVA 162
H + G+ WS +G+ + G G I Y+V+
Sbjct 144 IHKIITHEGLDAATVAGWSTKGKYIIAGHKDGKISKYDVS 183
> ath:AT5G67320 HOS15; HOS15 (high expression of osmotically responsive
genes 15)
Length=613
Score = 49.3 bits (116), Expect = 6e-06, Method: Composition-based stats.
Identities = 38/128 (29%), Positives = 57/128 (44%), Gaps = 13/128 (10%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGS----- 57
V+CV++ P L+ C D K+W++ +L T +YT+ SP G
Sbjct 451 VNCVKWDP--TGSLLASCSDDSTAKIWNIKQSTFVHDLREHTKEIYTIRWSPTGPGTNNP 508
Query 58 ----LCAFGGKDGVTILWDLSEGKHLFSLDGSC-PINSLGFFP-CKYWVGGATDKFFKIW 111
A D LWD GK L S +G P+ SL F P +Y G+ DK IW
Sbjct 509 NKQLTLASASFDSTVKLWDAELGKMLCSFNGHREPVYSLAFSPNGEYIASGSLDKSIHIW 568
Query 112 APEKKKVL 119
+ ++ K++
Sbjct 569 SIKEGKIV 576
> xla:444261 apaf1, MGC80868, apaf-1; apoptotic peptidase activating
factor 1; K02084 apoptotic protease-activating factor
Length=1248
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 42/164 (25%), Positives = 70/164 (42%), Gaps = 7/164 (4%)
Query 6 VRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKD 65
V+FF +E +F +D VKVW L T KL + + + + +SPD + + D
Sbjct 1046 VKFFKILQESQVFSWSFDGTVKVWDLITGKLRKEFICHSETVLSCDISPDSTKFSTASAD 1105
Query 66 GVTILWDLSEGKHLFSLDG--SCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLD--- 120
+W L L L+G SC + KY G + IW+ + ++L
Sbjct 1106 KSAKIWSLDMSTLLHELNGHQSCVRCCRFSWDNKYLATGDDNGKIMIWSVQNGELLKQCC 1165
Query 121 DISPDHPVKRGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
DIS ++ W ++S + I S+G N+ ++V G
Sbjct 1166 DISVNNENSLHDGWVTDLHFSPNSK--LIVSSGANVKWWDVDTG 1207
Score = 38.9 bits (89), Expect = 0.008, Method: Composition-based stats.
Identities = 45/191 (23%), Positives = 70/191 (36%), Gaps = 54/191 (28%)
Query 2 WVSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPA--NLV---------GQTFVLYT- 49
WV CV+F P K D+ VK+W + P+ NL +T VL T
Sbjct 880 WVHCVKFSP--KSSSFLTSSDDQTVKLWETSNVSKPSATNLKREFDVSFNGEETLVLATS 937
Query 50 ----------------------------VTVSPDGSLCAFGGKDGVTILWDLSEGKHLFS 81
++ D L A G +DG + D+S G+ L
Sbjct 938 KDDCILLINGMTGETLSQINTQDKCVTCCCLTNDYQLAAIGDRDGKVKVIDVSRGEILCK 997
Query 82 LDG------SCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLDDISPDHPVK--RGIP 133
LDG C + G K+ V + D ++W+ + L+ PVK + +
Sbjct 998 LDGHSATVQHCQFTADG----KHLVSSSDDSTIRVWSLASGESLELRGHKEPVKFFKILQ 1053
Query 134 WGPFFNWSYEG 144
F+WS++G
Sbjct 1054 ESQVFSWSFDG 1064
Score = 38.5 bits (88), Expect = 0.010, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 33/81 (40%), Gaps = 0/81 (0%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
V+C +F PL+ C D + +W T L+G T +Y SPD A
Sbjct 702 VNCCQFTNGLSAPLLATCSNDCFIMLWDSETEYSRNTLIGHTGAVYHCRYSPDDRYLASS 761
Query 63 GKDGVTILWDLSEGKHLFSLD 83
DG +WD+ S++
Sbjct 762 SMDGSLKIWDVESANEEKSIE 782
> sce:YPL151C PRP46, NTC50; Member of the NineTeen Complex (NTC)
that contains Prp19p and stabilizes U6 snRNA in catalytic
forms of the spliceosome containing U2, U5, and U6 snRNAs;
K12862 pleiotropic regulator 1
Length=451
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/101 (29%), Positives = 46/101 (45%), Gaps = 2/101 (1%)
Query 13 KEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWD 72
+ P +F DK VK W L ++ + G + TV++ P L A G+D V LWD
Sbjct 193 RHPYLFSVSEDKTVKCWDLEKNQIIRDYYGHLSGVRTVSIHPTLDLIATAGRDSVIKLWD 252
Query 73 LSEGKHLFSLDGSC-PINSLGFFPCK-YWVGGATDKFFKIW 111
+ + +L G PIN + P V +TD ++W
Sbjct 253 MRTRIPVITLVGHKGPINQVQCTPVDPQVVSSSTDATVRLW 293
Score = 36.2 bits (82), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 35/155 (22%), Positives = 55/155 (35%), Gaps = 7/155 (4%)
Query 11 FPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTIL 70
P LI G D ++K+W + T LVG + V +P D L
Sbjct 233 HPTLDLIATAGRDSVIKLWDMRTRIPVITLVGHKGPINQVQCTPVDPQVVSSSTDATVRL 292
Query 71 WDLSEGKHLFSL-DGSCPINSLGFFPCKYWVGGATDKFFKIWAPEKKKVLDDISPDHPVK 129
WD+ GK + L + + P ++ V A + W + +L + + K
Sbjct 293 WDVVAGKTMKVLTHHKRSVRATALHPKEFSVASACTDDIRSWGLAEGSLLTNFESE---K 349
Query 130 RGIPWGPFFNWSYEGRPLFIGSAGGNIPVYEVAEG 164
GI N + LF G G + Y+ G
Sbjct 350 TGIINTLSIN---QDDVLFAGGDNGVLSFYDYKSG 381
> dre:564019 tle1; transducin-like enhancer of split 1 (E(sp1)
homolog, Drosophila)
Length=826
Score = 48.5 bits (114), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 632 ACYALAISPDNKVCFSCCSDGNIVVWDLHNQTLVRQFQGHTDGASCIDISNDGTKLWTGG 691
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 692 LDNTVRCWDLREGRQLQQHDFTSQIFSLGYCPTGEWLA 729
Score = 34.7 bits (78), Expect = 0.16, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 43/109 (39%), Gaps = 12/109 (11%)
Query 11 FPKEPLIFLCGWDKLVKVWSLTTC--KLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVT 68
P + + G + +W L T ++ A L Y + +SPD +C DG
Sbjct 595 LPDGRTLIVGGEASTLSIWDLATPTPRIKAELTSSAPACYALAISPDNKVCFSCCSDGNI 654
Query 69 ILWDLSEGKHLFSLDG-----SC-PINSLGFFPCKYWVGGATDKFFKIW 111
++WDL + G SC I++ G K W GG D + W
Sbjct 655 VVWDLHNQTLVRQFQGHTDGASCIDISNDG---TKLWTGG-LDNTVRCW 699
> mmu:74187 Katnb1, 2410003J24Rik, KAT; katanin p80 (WD40-containing)
subunit B 1
Length=658
Score = 48.5 bits (114), Expect = 1e-05, Method: Composition-based stats.
Identities = 31/116 (26%), Positives = 53/116 (45%), Gaps = 2/116 (1%)
Query 23 DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSL 82
D +K+W + G + + + SPDG A D LWDL+ GK +
Sbjct 126 DTNIKLWDIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTVKLWDLTAGKMMSEF 185
Query 83 DG-SCPINSLGFFPCKYWVG-GATDKFFKIWAPEKKKVLDDISPDHPVKRGIPWGP 136
G + P+N + F P +Y + G++D+ + W EK +V+ I + R + + P
Sbjct 186 PGHTGPVNVVEFHPNEYLLASGSSDRTIRFWDLEKFQVVSCIEGEPGPVRSVLFNP 241
Score = 42.4 bits (98), Expect = 6e-04, Method: Composition-based stats.
Identities = 34/116 (29%), Positives = 50/116 (43%), Gaps = 5/116 (4%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
V C+RF P K + D VK+W LT K+ + G T + V P+ L A G
Sbjct 150 VRCLRFSPDGK--WLASAADDHTVKLWDLTAGKMMSEFPGHTGPVNVVEFHPNEYLLASG 207
Query 63 GKDGVTILWDLSEGKHLFSLDGS-CPINSLGFFP--CKYWVGGATDKFFKIWAPEK 115
D WDL + + + ++G P+ S+ F P C + G W PE+
Sbjct 208 SSDRTIRFWDLEKFQVVSCIEGEPGPVRSVLFNPDGCCLYSGCQDSLRVYGWEPER 263
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 31/115 (26%), Positives = 47/115 (40%), Gaps = 3/115 (2%)
Query 14 EPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDL 73
E LI ++VW L K+ L+G + ++ P G A G +D LWD+
Sbjct 75 EELIVAGSQSGSIRVWDLEAAKILRTLMGHKANICSLDFHPYGEFVASGSQDTNIKLWDI 134
Query 74 SEGKHLFSLDG-SCPINSLGFFPCKYWVGGATDKF-FKIWAPEKKKVLDDISPDH 126
+F G S + L F P W+ A D K+W K++ + P H
Sbjct 135 RRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTVKLWDLTAGKMMSEF-PGH 188
Score = 37.4 bits (85), Expect = 0.020, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 44/98 (44%), Gaps = 2/98 (2%)
Query 21 GWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLF 80
G D V +WS+ +L G T + +V ++ L G + G +WDL K L
Sbjct 40 GDDCRVNLWSINKPNCIMSLTGHTSPVESVRLNTPEELIVAGSQSGSIRVWDLEAAKILR 99
Query 81 SLDG-SCPINSLGFFPCKYWVG-GATDKFFKIWAPEKK 116
+L G I SL F P +V G+ D K+W +K
Sbjct 100 TLMGHKANICSLDFHPYGEFVASGSQDTNIKLWDIRRK 137
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDG-SCPINSLGF-FPCKYWVGGATDKFFKIWAP 113
G L A GG D LW +++ + SL G + P+ S+ P + V G+ ++W
Sbjct 33 GRLLATGGDDCRVNLWSINKPNCIMSLTGHTSPVESVRLNTPEELIVAGSQSGSIRVWDL 92
Query 114 EKKKVL 119
E K+L
Sbjct 93 EAAKIL 98
> hsa:10300 KATNB1, KAT; katanin p80 (WD repeat containing) subunit
B 1
Length=655
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 31/116 (26%), Positives = 53/116 (45%), Gaps = 2/116 (1%)
Query 23 DKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSL 82
D +K+W + G + + + SPDG A D LWDL+ GK +
Sbjct 126 DTNIKLWDIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTVKLWDLTAGKMMSEF 185
Query 83 DG-SCPINSLGFFPCKYWVG-GATDKFFKIWAPEKKKVLDDISPDHPVKRGIPWGP 136
G + P+N + F P +Y + G++D+ + W EK +V+ I + R + + P
Sbjct 186 PGHTGPVNVVEFHPNEYLLASGSSDRTIRFWDLEKFQVVSCIEGEPGPVRSVLFNP 241
Score = 42.4 bits (98), Expect = 6e-04, Method: Composition-based stats.
Identities = 34/116 (29%), Positives = 50/116 (43%), Gaps = 5/116 (4%)
Query 3 VSCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFG 62
V C+RF P K + D VK+W LT K+ + G T + V P+ L A G
Sbjct 150 VRCLRFSPDGK--WLASAADDHTVKLWDLTAGKMMSEFPGHTGPVNVVEFHPNEYLLASG 207
Query 63 GKDGVTILWDLSEGKHLFSLDGS-CPINSLGFFP--CKYWVGGATDKFFKIWAPEK 115
D WDL + + + ++G P+ S+ F P C + G W PE+
Sbjct 208 SSDRTIRFWDLEKFQVVSCIEGEPGPVRSVLFNPDGCCLYSGCQDSLRVYGWEPER 263
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 31/115 (26%), Positives = 47/115 (40%), Gaps = 3/115 (2%)
Query 14 EPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDL 73
E LI ++VW L K+ L+G + ++ P G A G +D LWD+
Sbjct 75 EELIVAGSQSGSIRVWDLEAAKILRTLMGHKANICSLDFHPYGEFVASGSQDTNIKLWDI 134
Query 74 SEGKHLFSLDG-SCPINSLGFFPCKYWVGGATDKF-FKIWAPEKKKVLDDISPDH 126
+F G S + L F P W+ A D K+W K++ + P H
Sbjct 135 RRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTVKLWDLTAGKMMSEF-PGH 188
Score = 37.4 bits (85), Expect = 0.020, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 44/98 (44%), Gaps = 2/98 (2%)
Query 21 GWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLF 80
G D V +WS+ +L G T + +V ++ L G + G +WDL K L
Sbjct 40 GDDCRVNLWSINKPNCIMSLTGHTSPVESVRLNTPEELIVAGSQSGSIRVWDLEAAKILR 99
Query 81 SLDG-SCPINSLGFFPCKYWVG-GATDKFFKIWAPEKK 116
+L G I SL F P +V G+ D K+W +K
Sbjct 100 TLMGHKANICSLDFHPYGEFVASGSQDTNIKLWDIRRK 137
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query 56 GSLCAFGGKDGVTILWDLSEGKHLFSLDG-SCPINSLGF-FPCKYWVGGATDKFFKIWAP 113
G L A GG D LW +++ + SL G + P+ S+ P + V G+ ++W
Sbjct 33 GRLLATGGDDCRVNLWSINKPNCIMSLTGHTSPVESVRLNTPEELIVAGSQSGSIRVWDL 92
Query 114 EKKKVL 119
E K+L
Sbjct 93 EAAKIL 98
> xla:398072 tle2, esg, esg1, esg2, esg3, grg2, grg3, tle3, tle3a,
xgrg3; transducin-like enhancer of split 2 (E(sp1) homolog,
Drosophila)
Length=768
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 574 ACYALAISPDAKVCFSCCSDGNIVVWDLQNQTLVRQFQGHTDGASCIDISHDGTKLWTGG 633
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 634 LDNTVRCWDLREGRQLQQHDFNSQIFSLGYCPTGEWLA 671
Score = 34.3 bits (77), Expect = 0.22, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 38/92 (41%), Gaps = 8/92 (8%)
Query 26 VKVWSLT--TCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSL- 82
+ +W L T ++ A L Y + +SPD +C DG ++WDL +
Sbjct 552 LSIWDLASPTPRIKAELTSSAPACYALAISPDAKVCFSCCSDGNIVVWDLQNQTLVRQFQ 611
Query 83 ---DGSCPINSLGFFPCKYWVGGATDKFFKIW 111
DG+ I+ + K W GG D + W
Sbjct 612 GHTDGASCID-ISHDGTKLWTGG-LDNTVRCW 641
> xla:779361 tle1, esg, esg1, grg1, tle3b, xgrg1; transducin-like
enhancer of split 1 (E(sp1) homolog)
Length=756
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 562 ACYALAISPDAKVCFSCCSDGNIVVWDLQNQTLVRQFQGHTDGASCIDISHDGTKLWTGG 621
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 622 LDNTVRCWDLREGRQLQQHDFNSQIFSLGYCPTGEWLA 659
Score = 33.9 bits (76), Expect = 0.22, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 38/92 (41%), Gaps = 8/92 (8%)
Query 26 VKVWSLT--TCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSEGKHLFSL- 82
+ +W L T ++ A L Y + +SPD +C DG ++WDL +
Sbjct 540 LSIWDLASPTPRIKAELTSSAPACYALAISPDAKVCFSCCSDGNIVVWDLQNQTLVRQFQ 599
Query 83 ---DGSCPINSLGFFPCKYWVGGATDKFFKIW 111
DG+ I+ + K W GG D + W
Sbjct 600 GHTDGASCID-ISHDGTKLWTGG-LDNTVRCW 629
> xla:399397 tle4, ESG, MGC86378, XGrg-4, XTCF-4, Xgrg4, Xtle4,
esg4, grg4; transducin-like enhancer of split 4 (E(sp1) homolog)
Length=772
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 578 ACYALAISPDSKVCFSCCSDGNIAVWDLHNQTLVRQFQGHTDGASCIDISNDGTKLWTGG 637
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 638 LDNTVRSWDLREGRQLQQHDFTSQIFSLGYCPTGEWLA 675
Score = 31.6 bits (70), Expect = 1.1, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 42/109 (38%), Gaps = 12/109 (11%)
Query 11 FPKEPLIFLCGWDKLVKVWSLT--TCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVT 68
P + + G + +W L T ++ A L Y + +SPD +C DG
Sbjct 541 LPDGRTLIVGGEASTLSIWDLAAPTPRIKAELTSSAPACYALAISPDSKVCFSCCSDGNI 600
Query 69 ILWDLSEGKHLFSLDG-----SC-PINSLGFFPCKYWVGGATDKFFKIW 111
+WDL + G SC I++ G K W GG D + W
Sbjct 601 AVWDLHNQTLVRQFQGHTDGASCIDISNDG---TKLWTGG-LDNTVRSW 645
> hsa:7091 TLE4, BCE-1, BCE1, E(spI), ESG, ESG4, GRG4; transducin-like
enhancer of split 4 (E(sp1) homolog, Drosophila)
Length=773
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 579 ACYALAISPDSKVCFSCCSDGNIAVWDLHNQTLVRQFQGHTDGASCIDISNDGTKLWTGG 638
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 639 LDNTVRSWDLREGRQLQQHDFTSQIFSLGYCPTGEWLA 676
Score = 31.6 bits (70), Expect = 1.2, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 42/109 (38%), Gaps = 12/109 (11%)
Query 11 FPKEPLIFLCGWDKLVKVWSLT--TCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVT 68
P + + G + +W L T ++ A L Y + +SPD +C DG
Sbjct 542 LPDGRTLIVGGEASTLSIWDLAAPTPRIKAELTSSAPACYALAISPDSKVCFSCCSDGNI 601
Query 69 ILWDLSEGKHLFSLDG-----SC-PINSLGFFPCKYWVGGATDKFFKIW 111
+WDL + G SC I++ G K W GG D + W
Sbjct 602 AVWDLHNQTLVRQFQGHTDGASCIDISNDG---TKLWTGG-LDNTVRSW 646
> mmu:21888 Tle4, 5730411M05Rik, AA792082, Bce-1, Bce1, ESTM13,
ESTM14, Grg4, X83332, X83333; transducin-like enhancer of
split 4, homolog of Drosophila E(spl)
Length=773
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 579 ACYALAISPDSKVCFSCCSDGNIAVWDLHNQTLVRQFQGHTDGASCIDISNDGTKLWTGG 638
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 639 LDNTVRSWDLREGRQLQQHDFTSQIFSLGYCPTGEWLA 676
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 42/109 (38%), Gaps = 12/109 (11%)
Query 11 FPKEPLIFLCGWDKLVKVWSLT--TCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVT 68
P + + G + +W L T ++ A L Y + +SPD +C DG
Sbjct 542 LPDGRTLIVGGEASTLSIWDLAAPTPRIKAELTSSAPACYALAISPDSKVCFSCCSDGNI 601
Query 69 ILWDLSEGKHLFSLDG-----SC-PINSLGFFPCKYWVGGATDKFFKIW 111
+WDL + G SC I++ G K W GG D + W
Sbjct 602 AVWDLHNQTLVRQFQGHTDGASCIDISNDG---TKLWTGG-LDNTVRSW 646
> hsa:7090 TLE3, ESG, ESG3, FLJ39460, GRG3, HsT18976, KIAA1547;
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
Length=769
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 575 ACYALAISPDAKVCFSCCSDGNIAVWDLHNQTLVRQFQGHTDGASCIDISHDGTKLWTGG 634
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 635 LDNTVRSWDLREGRQLQQHDFTSQIFSLGYCPTGEWLA 672
Score = 31.6 bits (70), Expect = 1.2, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 27/65 (41%), Gaps = 2/65 (3%)
Query 11 FPKEPLIFLCGWDKLVKVWSLT--TCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVT 68
P + + G + +W L T ++ A L Y + +SPD +C DG
Sbjct 538 LPDGRTLIVGGEASTLTIWDLASPTPRIKAELTSSAPACYALAISPDAKVCFSCCSDGNI 597
Query 69 ILWDL 73
+WDL
Sbjct 598 AVWDL 602
> mmu:21887 Tle3, 2610103N05Rik, ESG, Grg3a, Grg3b, mKIAA1547;
transducin-like enhancer of split 3, homolog of Drosophila
E(spl)
Length=782
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 40/98 (40%), Gaps = 0/98 (0%)
Query 4 SCVRFFPFPKEPLIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGG 63
+C P + F C D + VW L L G T + +S DG+ GG
Sbjct 588 ACYALAISPDAKVCFSCCSDGNIAVWDLHNQTLVRQFQGHTDGASCIDISHDGTKLWTGG 647
Query 64 KDGVTILWDLSEGKHLFSLDGSCPINSLGFFPCKYWVG 101
D WDL EG+ L D + I SLG+ P W+
Sbjct 648 LDNTVRSWDLREGRQLQQHDFTSQIFSLGYCPTGEWLA 685
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 27/65 (41%), Gaps = 2/65 (3%)
Query 11 FPKEPLIFLCGWDKLVKVWSLT--TCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVT 68
P + + G + +W L T ++ A L Y + +SPD +C DG
Sbjct 551 LPDGRTLIVGGEASTLTIWDLASPTPRIKAELTSSAPACYALAISPDAKVCFSCCSDGNI 610
Query 69 ILWDL 73
+WDL
Sbjct 611 AVWDL 615
> cel:K04G11.4 hypothetical protein
Length=395
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 56/124 (45%), Gaps = 3/124 (2%)
Query 16 LIFLCGWDKLVKVWSLTTCKLPANLVGQTFVLYTVTVSPDGSLCAFGGKDGVTILWDLSE 75
LI C DKLVKV+ +++ + L G T ++ +P G+L A G D +W
Sbjct 162 LIVSCSDDKLVKVFDVSSGRCVKTLKGHTNYVFCCCFNPSGTLIASGSFDETIRIWCARN 221
Query 76 GKHLFSLDG-SCPINSLGF-FPCKYWVGGATDKFFKIWAPEKKKVLDD-ISPDHPVKRGI 132
G +FS+ G P++S+ F Y G+ D +IW + I +HP +
Sbjct 222 GNTIFSIPGHEDPVSSVCFNRDGAYLASGSYDGIVRIWDSTTGTCVKTLIDEEHPPITHV 281
Query 133 PWGP 136
+ P
Sbjct 282 KFSP 285
Lambda K H
0.324 0.145 0.512
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3897828240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40