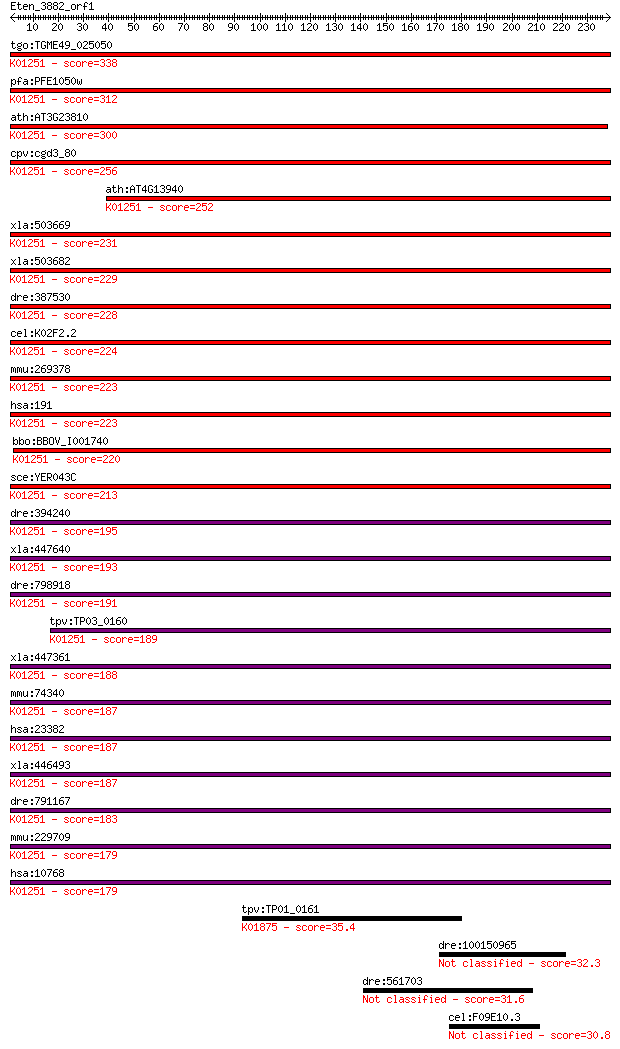
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3882_orf1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_025050 adenosylhomocysteinase, putative (EC:3.3.1.1... 338 1e-92
pfa:PFE1050w adenosylhomocysteinase(S-adenosyl-L-homocystein e... 312 6e-85
ath:AT3G23810 SAHH2; SAHH2 (S-ADENOSYL-L-HOMOCYSTEINE (SAH) HY... 300 4e-81
cpv:cgd3_80 S-adenosylhomocysteinase ; K01251 adenosylhomocyst... 256 5e-68
ath:AT4G13940 MEE58; MEE58 (MATERNAL EFFECT EMBRYO ARREST 58);... 252 7e-67
xla:503669 ahcy-a, sahh; adenosylhomocysteinase (EC:3.3.1.1); ... 231 2e-60
xla:503682 ahcy-b, ahcy, sahh; adenosylhomocysteinase (EC:3.3.... 229 5e-60
dre:387530 ahcy, D150, cb1079, hm:zeh1173, hm:zeh1364, wu:fj67... 228 1e-59
cel:K02F2.2 hypothetical protein; K01251 adenosylhomocysteinas... 224 3e-58
mmu:269378 Ahcy, AA987153, AL024110, CuBP, MGC102079, MGC11806... 223 4e-58
hsa:191 AHCY, SAHH; adenosylhomocysteinase (EC:3.3.1.1); K0125... 223 7e-58
bbo:BBOV_I001740 19.m02133; adenosylhomocysteinase (S-adenosyl... 220 4e-57
sce:YER043C SAH1; S-adenosyl-L-homocysteine hydrolase, catabol... 213 4e-55
dre:394240 ahcyl2; S-adenosylhomocysteine hydrolase-like 2 (EC... 195 1e-49
xla:447640 ahcyl1, MGC86404, irbit; adenosylhomocysteinase-lik... 193 6e-49
dre:798918 ahcyl1, MGC64088, cb586, wu:fj66f02; S-adenosylhomo... 191 2e-48
tpv:TP03_0160 S-adenosyl-L-homocysteine hydrolase (EC:3.3.1.1)... 189 9e-48
xla:447361 ahcyl2, MGC84148; adenosylhomocysteinase-like 2 (EC... 188 1e-47
mmu:74340 Ahcyl2, 4631427C17Rik, AI227036, Adohcyase3, KIAA082... 187 2e-47
hsa:23382 AHCYL2, ADOHCYASE3, FLJ21719, KIAA0828; adenosylhomo... 187 3e-47
xla:446493 MGC79134 protein; K01251 adenosylhomocysteinase [EC... 187 4e-47
dre:791167 zgc:158222 (EC:3.3.1.1); K01251 adenosylhomocystein... 183 4e-46
mmu:229709 Ahcyl1, 1110034F20Rik, AA409031, AA414901, Ahcy-rs3... 179 9e-45
hsa:10768 AHCYL1, DCAL, IRBIT, PRO0233, XPVKONA; adenosylhomoc... 179 9e-45
tpv:TP01_0161 seryl-tRNA synthetase; K01875 seryl-tRNA synthet... 35.4 0.18
dre:100150965 retinol dehydrogenase 12-like 32.3 1.6
dre:561703 rictora, si:ch211-14b14.3; rapamycin-insensitive co... 31.6 2.5
cel:F09E10.3 dhs-25; DeHydrogenases, Short chain family member... 30.8 4.3
> tgo:TGME49_025050 adenosylhomocysteinase, putative (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=477
Score = 338 bits (867), Expect = 1e-92, Method: Compositional matrix adjust.
Identities = 157/238 (65%), Positives = 190/238 (79%), Gaps = 0/238 (0%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAI SA VFAWKGET+EEYWWC+ ALTW + AGPDLLVDDGGD T
Sbjct 77 CNIFSTQDHAAAAIVKNNSAAVFAWKGETLEEYWWCTVMALTWGEGAGPDLLVDDGGDAT 136
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
+LIHEGV AE+ + SG LP+ +A + E V ++L + P+KW ++AA ++GVSE
Sbjct 137 LLIHEGVKAEKEFARSGALPDPAATDNLEMKVVYKILAEGLQKDPQKWSRMAANLKGVSE 196
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV RLL+ A LLFPA+NVNDCVTKSKFDNVYGCRHSLPD IMRATDVMLGGK
Sbjct 197 ETTTGVHRLLQMAKKKELLFPAINVNDCVTKSKFDNVYGCRHSLPDGIMRATDVMLGGKR 256
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
V+CG+GDVGKGCAAA++G G RV+++E+DPICALQAAMEG+ V L++ + AD+F+
Sbjct 257 VVICGFGDVGKGCAAAMKGAGARVVVTEVDPICALQAAMEGYSVSTLESQLATADIFI 314
> pfa:PFE1050w adenosylhomocysteinase(S-adenosyl-L-homocystein
e hydrolase) (EC:3.3.1.1); K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=479
Score = 312 bits (800), Expect = 6e-85, Method: Compositional matrix adjust.
Identities = 140/240 (58%), Positives = 178/240 (74%), Gaps = 2/240 (0%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDC--AGPDLLVDDGGD 58
CNI+ST D+AAAA++ + TVFAWK ET+EEYWWC E ALTW D GPD++VDDGGD
Sbjct 78 CNIYSTADYAAAAVSTLENVTVFAWKNETLEEYWWCVESALTWGDGDDNGPDMIVDDGGD 137
Query 59 ITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGV 118
T+L+H+GV E++YE +LP+ A +EE LL+ S+ P+KW +A + GV
Sbjct 138 ATLLVHKGVEYEKLYEEKNILPDPEKAKNEEERCFLTLLKNSILKNPKKWTNIAKKIIGV 197
Query 119 SEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGG 178
SEETTTGVLRL + N LLF A+NVND VTK K+DNVYGCRHSLPD +MRATD ++ G
Sbjct 198 SEETTTGVLRLKKMDKQNELLFTAINVNDAVTKQKYDNVYGCRHSLPDGLMRATDFLISG 257
Query 179 KLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
K+ V+CGYGDVGKGCA++++G G RV I+EIDPICA+QA MEGF V+ LD +V D F+
Sbjct 258 KIVVICGYGDVGKGCASSMKGLGARVYITEIDPICAIQAVMEGFNVVTLDEIVDKGDFFI 317
> ath:AT3G23810 SAHH2; SAHH2 (S-ADENOSYL-L-HOMOCYSTEINE (SAH)
HYDROLASE 2); adenosylhomocysteinase/ binding / catalytic (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=485
Score = 300 bits (767), Expect = 4e-81, Method: Compositional matrix adjust.
Identities = 147/237 (62%), Positives = 183/237 (77%), Gaps = 1/237 (0%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA + SA VFAWKGET++EYWWC+E+AL W GPDL+VDDGGD T
Sbjct 86 CNIFSTQDHAAAAIARD-SAAVFAWKGETLQEYWWCTERALDWGPGGGPDLIVDDGGDAT 144
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
+LIHEGV AEE++ +G P+ ++ + E V +++ + P+K+ ++ + GVSE
Sbjct 145 LLIHEGVKAEEIFAKNGTFPDPTSTDNPEFQIVLSIIKDGLQVDPKKYHKMKERLVGVSE 204
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV RL + LLFPA+NVND VTKSKFDN+YGCRHSLPD +MRATDVM+ GK+
Sbjct 205 ETTTGVKRLYQMQETGALLFPAINVNDSVTKSKFDNLYGCRHSLPDGLMRATDVMIAGKV 264
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
AV+CGYGDVGKGCAAA++ G RVI++EIDPICALQA MEG VL L+ VV AD+F
Sbjct 265 AVICGYGDVGKGCAAAMKTAGARVIVTEIDPICALQALMEGLQVLTLEDVVSEADIF 321
> cpv:cgd3_80 S-adenosylhomocysteinase ; K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=495
Score = 256 bits (654), Expect = 5e-68, Method: Compositional matrix adjust.
Identities = 130/252 (51%), Positives = 163/252 (64%), Gaps = 14/252 (5%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPD------CAGPDLLVD 54
CNI+STQDHAAAA+ + ATVFAWK ET+E+YW C A+TW + GP+L+VD
Sbjct 77 CNIYSTQDHAAAALVKKNIATVFAWKNETIEDYWVCLNDAMTWRNPNDKDKICGPNLIVD 136
Query 55 DGGDITMLIHEGVHAEEVYESSGVLPE-VSAAADEEAAAVQ-------QLLRRSVAAAPR 106
DGGD T+++HEGV AE YE +PE + DE + ++L+ + P
Sbjct 137 DGGDATLILHEGVKAEIEYEKYNKIPEYLETELDENGKQLSMDLKCMYKVLKMELLKNPF 196
Query 107 KWRQLAAGVRGVSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPD 166
+WR + + GVSEETTTGVLRL + LL PA+NVND VTKSKFDN YGCR SL
Sbjct 197 RWRGMLKDLYGVSEETTTGVLRLKIMESEGKLLLPAINVNDSVTKSKFDNTYGCRQSLLH 256
Query 167 AIMRATDVMLGGKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLP 226
+ ML GK VV GYG+VGKGCA L G G RVI++EIDPICALQA+MEG+ V
Sbjct 257 GLFNGCIQMLAGKKIVVLGYGEVGKGCAQGLSGVGARVIVTEIDPICALQASMEGYQVSV 316
Query 227 LDAVVGVADLFV 238
L+ VV AD+F+
Sbjct 317 LEDVVSEADIFI 328
> ath:AT4G13940 MEE58; MEE58 (MATERNAL EFFECT EMBRYO ARREST 58);
adenosylhomocysteinase; K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=364
Score = 252 bits (644), Expect = 7e-67, Method: Compositional matrix adjust.
Identities = 118/200 (59%), Positives = 151/200 (75%), Gaps = 0/200 (0%)
Query 39 QALTWPDCAGPDLLVDDGGDITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLR 98
+AL W GPDL+VDDGGD T+LIHEGV AEE++E +G +P+ ++ + E V +++
Sbjct 2 RALDWGPGGGPDLIVDDGGDATLLIHEGVKAEEIFEKTGQVPDPTSTDNPEFQIVLSIIK 61
Query 99 RSVAAAPRKWRQLAAGVRGVSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVY 158
+ P+K+ ++ + GVSEETTTGV RL + LLFPA+NVND VTKSKFDN+Y
Sbjct 62 EGLQVDPKKYHKMKERLVGVSEETTTGVKRLYQMQQNGTLLFPAINVNDSVTKSKFDNLY 121
Query 159 GCRHSLPDAIMRATDVMLGGKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAA 218
GCRHSLPD +MRATDVM+ GK+AV+CGYGDVGKGCAAA++ G RVI++EIDPICALQA
Sbjct 122 GCRHSLPDGLMRATDVMIAGKVAVICGYGDVGKGCAAAMKTAGARVIVTEIDPICALQAL 181
Query 219 MEGFGVLPLDAVVGVADLFV 238
MEG VL L+ VV AD+FV
Sbjct 182 MEGLQVLTLEDVVSEADIFV 201
> xla:503669 ahcy-a, sahh; adenosylhomocysteinase (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=433
Score = 231 bits (588), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 122/238 (51%), Positives = 147/238 (61%), Gaps = 42/238 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA G V+AWKGET EEY WC EQ + + D ++++DDGGD+T
Sbjct 79 CNIFSTQDHAAAAIAKTG-VPVYAWKGETDEEYIWCIEQTIYFKDGKPLNMILDDGGDLT 137
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+H K+ QL G++G+SE
Sbjct 138 NLVHS-----------------------------------------KYPQLLKGIKGISE 156
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV L + ++ L PA+NVND VTKSKFDN+YGCR SL D I RATDVM+ GK+
Sbjct 157 ETTTGVHNLYKMKSSGTLQVPAINVNDSVTKSKFDNLYGCRESLIDGIKRATDVMIAGKV 216
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
AVV GYGDVGKGCA ALR G RVII+EIDPI ALQAAMEG+ V +D ++FV
Sbjct 217 AVVAGYGDVGKGCAQALRAFGARVIITEIDPINALQAAMEGYEVTTMDEASKEGNIFV 274
> xla:503682 ahcy-b, ahcy, sahh; adenosylhomocysteinase (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=433
Score = 229 bits (585), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 121/238 (50%), Positives = 147/238 (61%), Gaps = 42/238 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA G V+AWKGET EEY WC EQ + + D ++++DDGGD+T
Sbjct 79 CNIFSTQDHAAAAIAKTG-VPVYAWKGETDEEYIWCIEQTIYFKDGKPLNMILDDGGDLT 137
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+H K+ QL G++G+SE
Sbjct 138 NLVH-----------------------------------------TKYPQLLKGIKGISE 156
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV L + ++ L PA+NVND VTKSKFDN+YGCR SL D I RATDVM+ GK+
Sbjct 157 ETTTGVHNLYKMKSSGTLQVPAINVNDSVTKSKFDNLYGCRESLIDGIKRATDVMIAGKV 216
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
AVV GYGDVGKGCA ALR G RV+I+EIDPI ALQAAMEG+ V +D ++FV
Sbjct 217 AVVAGYGDVGKGCAQALRAFGARVLITEIDPINALQAAMEGYEVTTMDEASKEGNIFV 274
> dre:387530 ahcy, D150, cb1079, hm:zeh1173, hm:zeh1364, wu:fj67b02;
S-adenosylhomocysteine hydrolase (EC:3.3.1.1); K01251
adenosylhomocysteinase [EC:3.3.1.1]
Length=433
Score = 228 bits (582), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 121/238 (50%), Positives = 148/238 (62%), Gaps = 42/238 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA G V+AWKGET EEY WC EQ + + D ++++DDGGD+T
Sbjct 79 CNIFSTQDHAAAAIAKTG-VPVYAWKGETDEEYVWCIEQTIYFKDGQPLNMILDDGGDLT 137
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+H+ K+ +L AG++G+SE
Sbjct 138 NLVHQ-----------------------------------------KYPKLLAGIKGISE 156
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV L + + L PA+NVND VTKSKFDN+YGCR SL D I RATDVM+ GK+
Sbjct 157 ETTTGVHNLYKMLKSGELKVPAINVNDSVTKSKFDNLYGCRESLIDGIKRATDVMIAGKV 216
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
AVV GYGDVGKGC ALRG G RVI++EIDPI ALQAAMEG+ V +D ++FV
Sbjct 217 AVVAGYGDVGKGCVQALRGFGARVIVTEIDPINALQAAMEGYEVTTMDEACKEGNIFV 274
> cel:K02F2.2 hypothetical protein; K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=437
Score = 224 bits (570), Expect = 3e-58, Method: Compositional matrix adjust.
Identities = 122/238 (51%), Positives = 146/238 (61%), Gaps = 42/238 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA G V+AWKGET EEY WC EQ + + D ++++DDGGD+T
Sbjct 80 CNIFSTQDHAAAAIAQTG-VPVYAWKGETDEEYEWCIEQTIVFKDGQPLNMILDDGGDLT 138
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+H K+ Q AG+RG+SE
Sbjct 139 NLVHA-----------------------------------------KYPQYLAGIRGLSE 157
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV L + A L PA+NVND VTKSKFDN+YG R SLPD I RATDVML GK+
Sbjct 158 ETTTGVHNLAKMLAKGDLKVPAINVNDSVTKSKFDNLYGIRESLPDGIKRATDVMLAGKV 217
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
AVV GYGDVGKG AA+L+ G RVI++EIDPI ALQAAMEG+ V L+ A++ V
Sbjct 218 AVVAGYGDVGKGSAASLKAFGSRVIVTEIDPINALQAAMEGYEVTTLEEAAPKANIIV 275
> mmu:269378 Ahcy, AA987153, AL024110, CuBP, MGC102079, MGC118063,
MGC19228, SAHH; S-adenosylhomocysteine hydrolase (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=432
Score = 223 bits (569), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 129/239 (53%), Positives = 151/239 (63%), Gaps = 45/239 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGP-DLLVDDGGDI 59
CNIFSTQDHAAAAIA G VFAWKGET EEY WC EQ L + D GP ++++DDGGD+
Sbjct 79 CNIFSTQDHAAAAIAKAG-IPVFAWKGETDEEYLWCIEQTLHFKD--GPLNMILDDGGDL 135
Query 60 TMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVS 119
T LIH K+ QL +G+RG+S
Sbjct 136 TNLIHT-----------------------------------------KYPQLLSGIRGIS 154
Query 120 EETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGK 179
EETTTGV L + + +L PA+NVND VTKSKFDN+YGCR SL D I RATDVM+ GK
Sbjct 155 EETTTGVHNLYKMMSNGILKVPAINVNDSVTKSKFDNLYGCRESLIDGIKRATDVMIAGK 214
Query 180 LAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
+AVV GYGDVGKGCA ALRG G RVII+EIDPI ALQAAMEG+ V +D ++FV
Sbjct 215 VAVVAGYGDVGKGCAQALRGFGARVIITEIDPINALQAAMEGYEVTTMDEACKEGNIFV 273
> hsa:191 AHCY, SAHH; adenosylhomocysteinase (EC:3.3.1.1); K01251
adenosylhomocysteinase [EC:3.3.1.1]
Length=432
Score = 223 bits (567), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 129/239 (53%), Positives = 150/239 (62%), Gaps = 45/239 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGP-DLLVDDGGDI 59
CNIFSTQDHAAAAIA G V+AWKGET EEY WC EQ L + D GP ++++DDGGD+
Sbjct 79 CNIFSTQDHAAAAIAKAG-IPVYAWKGETDEEYLWCIEQTLYFKD--GPLNMILDDGGDL 135
Query 60 TMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVS 119
T LIH K+ QL G+RG+S
Sbjct 136 TNLIHT-----------------------------------------KYPQLLPGIRGIS 154
Query 120 EETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGK 179
EETTTGV L + A +L PA+NVND VTKSKFDN+YGCR SL D I RATDVM+ GK
Sbjct 155 EETTTGVHNLYKMMANGILKVPAINVNDSVTKSKFDNLYGCRESLIDGIKRATDVMIAGK 214
Query 180 LAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
+AVV GYGDVGKGCA ALRG G RVII+EIDPI ALQAAMEG+ V +D ++FV
Sbjct 215 VAVVAGYGDVGKGCAQALRGFGARVIITEIDPINALQAAMEGYEVTTMDEACQEGNIFV 273
> bbo:BBOV_I001740 19.m02133; adenosylhomocysteinase (S-adenosyl-L-homocysteinehydrolase)
(EC:3.3.1.1); K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=491
Score = 220 bits (560), Expect = 4e-57, Method: Compositional matrix adjust.
Identities = 116/242 (47%), Positives = 144/242 (59%), Gaps = 5/242 (2%)
Query 2 NIFSTQDHAAAAIAA--EGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDI 59
N FS D AA+ A T+FAWKGETVEEYWWC Q+L WP+ GP L+VDDG
Sbjct 87 NPFSAHDGICAALKAFHHDETTIFAWKGETVEEYWWCVYQSLRWPNADGPQLIVDDGCSA 146
Query 60 TMLIHEGVHAEEVYESSGVL--PEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
+I G+ E + G L P+ D + + L V W +LA V G
Sbjct 147 YTMIKYGLDLERRHAKDGSLLNPKDFDENDRDIQCLVTFLNTLVTKEGYFWEKLANQVVG 206
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATD-VML 176
+SEETT+GV + A LLFP M+ NDCVTK K+DN+YGCRHS D ++
Sbjct 207 LSEETTSGVTHFRKFWRAEGLLFPVMSTNDCVTKQKYDNIYGCRHSGIHGFFNGGDGFLI 266
Query 177 GGKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADL 236
GGK VV GYG+VGKG A RGQG +V I+EIDPICALQAAMEGF V+ L+ V+ AD+
Sbjct 267 GGKTVVVIGYGNVGKGVAQGFRGQGAKVKITEIDPICALQAAMEGFDVVLLEDVLETADI 326
Query 237 FV 238
F+
Sbjct 327 FI 328
> sce:YER043C SAH1; S-adenosyl-L-homocysteine hydrolase, catabolizes
S-adenosyl-L-homocysteine which is formed after donation
of the activated methyl group of S-adenosyl-L-methionine
(AdoMet) to an acceptor (EC:3.3.1.1); K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=449
Score = 213 bits (543), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 124/239 (51%), Positives = 146/239 (61%), Gaps = 43/239 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQAL-TWPDCAGPDLLVDDGGDI 59
CNI+STQDHAAAAIAA G VFAWKGET EEY WC EQ L + D +L++DDGGD+
Sbjct 80 CNIYSTQDHAAAAIAASG-VPVFAWKGETEEEYLWCIEQQLFAFKDNKKLNLILDDGGDL 138
Query 60 TMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVS 119
T L+HE K ++ G+S
Sbjct 139 TTLVHE-----------------------------------------KHPEMLEDCFGLS 157
Query 120 EETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGK 179
EETTTGV L R L PA+NVND VTKSKFDN+YGCR SL D I RATDVML GK
Sbjct 158 EETTTGVHHLYRMVKEGKLKVPAINVNDSVTKSKFDNLYGCRESLVDGIKRATDVMLAGK 217
Query 180 LAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
+AVV GYGDVGKGCAAALRG G RV+++EIDPI ALQAAMEG+ V+ ++ + +FV
Sbjct 218 VAVVAGYGDVGKGCAAALRGMGARVLVTEIDPINALQAAMEGYQVVTMEDASHIGQVFV 276
> dre:394240 ahcyl2; S-adenosylhomocysteine hydrolase-like 2 (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=591
Score = 195 bits (495), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 101/241 (41%), Positives = 141/241 (58%), Gaps = 49/241 (20%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALT---WPDCAGPDLLVDDGG 57
CNI+STQ+ AAA+A EG +VFAWKGE+ +++WWC ++ + W P++++DDGG
Sbjct 238 CNIYSTQNEVAAALA-EGGFSVFAWKGESEDDFWWCIDRCVNVEGW----QPNMILDDGG 292
Query 58 DITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
D+T I+ +K+ + ++G
Sbjct 293 DLTHWIY-----------------------------------------KKYPNMFKKIKG 311
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
+ EE+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TDVM G
Sbjct 312 IVEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFG 371
Query 178 GKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
GK VVCGYG+VGKGC+AAL+ G V ++EIDPICALQA M+GF ++ L V+ D+
Sbjct 372 GKQVVVCGYGEVGKGCSAALKAMGSIVYVTEIDPICALQACMDGFRLVKLSEVIRQVDIV 431
Query 238 V 238
+
Sbjct 432 I 432
> xla:447640 ahcyl1, MGC86404, irbit; adenosylhomocysteinase-like
1 (EC:3.3.1.1); K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=520
Score = 193 bits (490), Expect = 6e-49, Method: Compositional matrix adjust.
Identities = 100/238 (42%), Positives = 138/238 (57%), Gaps = 42/238 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQ+ AAA+A G A +FAWKGE+ +++WWC ++ + ++++DDGGD+T
Sbjct 166 CNIFSTQNEVAAALAESGVA-IFAWKGESEDDFWWCIDRCVNADSGWQANMILDDGGDLT 224
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
++ +K+ + +RG+ E
Sbjct 225 HWVY-----------------------------------------KKYPHVYTQIRGIVE 243
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R+TDVM GGK
Sbjct 244 ESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRSTDVMFGGKQ 303
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VVCGYG+VGKGC AAL+ G V ++EIDPICALQA MEGF V+ L ++ D+ +
Sbjct 304 VVVCGYGEVGKGCCAALKALGAVVHVTEIDPICALQACMEGFRVVKLSEIIRQVDVVI 361
> dre:798918 ahcyl1, MGC64088, cb586, wu:fj66f02; S-adenosylhomocysteine
hydrolase-like 1 (EC:3.3.1.1); K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=512
Score = 191 bits (484), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 101/238 (42%), Positives = 139/238 (58%), Gaps = 43/238 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNI+STQ+ AAA+A G +VFAWKGE+ +++WWC ++ + D ++++DDGGD+T
Sbjct 159 CNIYSTQNEVAAALAESG-VSVFAWKGESEDDFWWCIDRCVNM-DGWQANMILDDGGDLT 216
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
++ +K+ + VRG+ E
Sbjct 217 HWVY-----------------------------------------KKYPSVFKKVRGIVE 235
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TDVM GGK
Sbjct 236 ESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQ 295
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VVCGYG+VGKGC +AL+ G V ++EIDPICALQA M+GF V+ L+ VV D+ +
Sbjct 296 VVVCGYGEVGKGCCSALKALGAIVCVTEIDPICALQACMDGFRVVKLNEVVRQMDMVI 353
> tpv:TP03_0160 S-adenosyl-L-homocysteine hydrolase (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=489
Score = 189 bits (479), Expect = 9e-48, Method: Compositional matrix adjust.
Identities = 97/225 (43%), Positives = 134/225 (59%), Gaps = 3/225 (1%)
Query 17 EGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDITMLIHEGVHAEEVYESS 76
+G T+F +KGET+EEY+WC Q+L WPD + P LLVDDG +IH G E++Y
Sbjct 104 KGKVTLFGYKGETIEEYFWCMYQSLNWPDTSMPLLLVDDGCCSYNMIHYGKRLEDMYREK 163
Query 77 GVLPEVSA--AADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSEETTTGVLRLLRRAA 134
G L ++S +D + A+ L V P + L G+SEETT+G+ + +
Sbjct 164 GQLYDLSVLEPSDNDTRALLNFLNHLVTKKPDFFTNLVKRTVGLSEETTSGLTEMRKLYK 223
Query 135 ANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATD-VMLGGKLAVVCGYGDVGKGC 193
++ LLFP + ND V K+KFDN YG R+S D + R + V+LGG+ VV GYG+ GKG
Sbjct 224 SHGLLFPIYSTNDVVCKTKFDNNYGARYSSIDGMHRGVEGVLLGGRQVVVVGYGNTGKGV 283
Query 194 AAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
RG G V + E DPICALQ M+G+ V+ L+ VV AD+FV
Sbjct 284 CMGFRGAGAIVKVCEADPICALQCVMDGYQVVLLEDVVETADIFV 328
> xla:447361 ahcyl2, MGC84148; adenosylhomocysteinase-like 2 (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=588
Score = 188 bits (478), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 98/241 (40%), Positives = 139/241 (57%), Gaps = 49/241 (20%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALT---WPDCAGPDLLVDDGG 57
CNI+ST + AAA+A G A VF+WKGE+ +++WWC ++ + W P++++DDGG
Sbjct 235 CNIYSTLNEVAAALAENGVA-VFSWKGESEDDFWWCIDRCVNVEGW----QPNMILDDGG 289
Query 58 DITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
D+T I+ +K+ + ++G
Sbjct 290 DLTHWIY-----------------------------------------KKYPNMFKKIKG 308
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
+ EE+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TD+M G
Sbjct 309 IVEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDMMFG 368
Query 178 GKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
GK VVCGYG+VGKGC AAL+ G V ++EIDPICALQA M+GF ++ L+ V+ D+
Sbjct 369 GKQVVVCGYGEVGKGCCAALKAMGSIVYVTEIDPICALQACMDGFRLVKLNEVIRQVDIV 428
Query 238 V 238
+
Sbjct 429 I 429
> mmu:74340 Ahcyl2, 4631427C17Rik, AI227036, Adohcyase3, KIAA0828,
mKIAA0828; S-adenosylhomocysteine hydrolase-like 2 (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=642
Score = 187 bits (476), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 98/241 (40%), Positives = 138/241 (57%), Gaps = 49/241 (20%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALT---WPDCAGPDLLVDDGG 57
CNI+ST + AAA+A G VFAWKGE+ +++WWC ++ + W P++++DDGG
Sbjct 289 CNIYSTLNEVAAALAESGFP-VFAWKGESEDDFWWCIDRCVNVEGW----QPNMILDDGG 343
Query 58 DITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
D+T I+ +K+ + ++G
Sbjct 344 DLTHWIY-----------------------------------------KKYPNMFKKIKG 362
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
+ EE+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TD+M G
Sbjct 363 IVEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDMMFG 422
Query 178 GKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
GK VVCGYG+VGKGC AAL+ G V ++EIDPICALQA M+GF ++ L+ V+ D+
Sbjct 423 GKQVVVCGYGEVGKGCCAALKAMGSIVYVTEIDPICALQACMDGFRLVKLNEVIRQVDIV 482
Query 238 V 238
+
Sbjct 483 I 483
> hsa:23382 AHCYL2, ADOHCYASE3, FLJ21719, KIAA0828; adenosylhomocysteinase-like
2 (EC:3.3.1.1); K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=610
Score = 187 bits (475), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 98/241 (40%), Positives = 138/241 (57%), Gaps = 49/241 (20%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALT---WPDCAGPDLLVDDGG 57
CNI+ST + AAA+A G VFAWKGE+ +++WWC ++ + W P++++DDGG
Sbjct 257 CNIYSTLNEVAAALAESGFP-VFAWKGESEDDFWWCIDRCVNVEGW----QPNMILDDGG 311
Query 58 DITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
D+T I+ +K+ + ++G
Sbjct 312 DLTHWIY-----------------------------------------KKYPNMFKKIKG 330
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
+ EE+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TD+M G
Sbjct 331 IVEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDMMFG 390
Query 178 GKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
GK VVCGYG+VGKGC AAL+ G V ++EIDPICALQA M+GF ++ L+ V+ D+
Sbjct 391 GKQVVVCGYGEVGKGCCAALKAMGSIVYVTEIDPICALQACMDGFRLVKLNEVIRQVDIV 450
Query 238 V 238
+
Sbjct 451 I 451
> xla:446493 MGC79134 protein; K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=583
Score = 187 bits (474), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 97/241 (40%), Positives = 138/241 (57%), Gaps = 49/241 (20%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALT---WPDCAGPDLLVDDGG 57
CNI+ST + AAA+A G VF+WKGE+ +++WWC ++ + W P++++DDGG
Sbjct 230 CNIYSTLNEVAAALAESG-VPVFSWKGESEDDFWWCIDRCVNVEGW----QPNMILDDGG 284
Query 58 DITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
D+T I+ +K+ + ++G
Sbjct 285 DLTHWIY-----------------------------------------KKYPNMFKKIKG 303
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
+ EE+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TD+M G
Sbjct 304 IVEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDMMFG 363
Query 178 GKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
GK VVCGYG+VGKGC AAL+ G V ++EIDPICALQA M+GF ++ L+ V+ D+
Sbjct 364 GKQVVVCGYGEVGKGCCAALKAMGSIVYVTEIDPICALQACMDGFRLVKLNEVIRQVDIV 423
Query 238 V 238
+
Sbjct 424 I 424
> dre:791167 zgc:158222 (EC:3.3.1.1); K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=497
Score = 183 bits (465), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 97/241 (40%), Positives = 137/241 (56%), Gaps = 49/241 (20%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALT---WPDCAGPDLLVDDGG 57
CNI+STQ+ AAA+A E VFAWKGE+ +++WWC ++ + W P++++DDGG
Sbjct 144 CNIYSTQNEVAAALA-ERDFPVFAWKGESEDDFWWCIDRCVNVEGWE----PNMILDDGG 198
Query 58 DITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
D+T I+ +K+ + V+G
Sbjct 199 DMTHWIY-----------------------------------------KKYPHIFKKVKG 217
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
+ EE+ TG+ RL + A L PA+NVND VTK KFDN+Y CR S+ D++ + D+M G
Sbjct 218 IVEESITGIHRLHHLSKAGKLCVPAINVNDSVTKQKFDNLYCCRESILDSLKKTADIMFG 277
Query 178 GKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
GK VVCGYG+VGKGC+AAL+ G V ++EIDPICALQA M+GF + L VV D+
Sbjct 278 GKQVVVCGYGEVGKGCSAALKAMGSIVYVTEIDPICALQACMDGFRLTKLSDVVRQVDMV 337
Query 238 V 238
+
Sbjct 338 I 338
> mmu:229709 Ahcyl1, 1110034F20Rik, AA409031, AA414901, Ahcy-rs3,
DCAL, Irbit; S-adenosylhomocysteine hydrolase-like 1 (EC:3.3.1.1);
K01251 adenosylhomocysteinase [EC:3.3.1.1]
Length=530
Score = 179 bits (453), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 100/238 (42%), Positives = 137/238 (57%), Gaps = 43/238 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNI+STQ+ AAA+A VFAWKGE+ +++WWC ++ + D ++++DDGGD+T
Sbjct 177 CNIYSTQNEVAAALAEA-GVAVFAWKGESEDDFWWCIDRCVNM-DGWQANMILDDGGDLT 234
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
++ +K+ + +RG+ E
Sbjct 235 HWVY-----------------------------------------KKYPNVFKKIRGIVE 253
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TDVM GGK
Sbjct 254 ESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQ 313
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VVCGYG+VGKGC AAL+ G V I+EIDPICALQA M+GF V+ L+ V+ D+ +
Sbjct 314 VVVCGYGEVGKGCCAALKALGAIVYITEIDPICALQACMDGFRVVKLNEVIRQVDVVI 371
> hsa:10768 AHCYL1, DCAL, IRBIT, PRO0233, XPVKONA; adenosylhomocysteinase-like
1 (EC:3.3.1.1); K01251 adenosylhomocysteinase
[EC:3.3.1.1]
Length=530
Score = 179 bits (453), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 100/238 (42%), Positives = 137/238 (57%), Gaps = 43/238 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNI+STQ+ AAA+A VFAWKGE+ +++WWC ++ + D ++++DDGGD+T
Sbjct 177 CNIYSTQNEVAAALAEA-GVAVFAWKGESEDDFWWCIDRCVNM-DGWQANMILDDGGDLT 234
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
++ +K+ + +RG+ E
Sbjct 235 HWVY-----------------------------------------KKYPNVFKKIRGIVE 253
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TDVM GGK
Sbjct 254 ESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQ 313
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VVCGYG+VGKGC AAL+ G V I+EIDPICALQA M+GF V+ L+ V+ D+ +
Sbjct 314 VVVCGYGEVGKGCCAALKALGAIVYITEIDPICALQACMDGFRVVKLNEVIRQVDVVI 371
> tpv:TP01_0161 seryl-tRNA synthetase; K01875 seryl-tRNA synthetase
[EC:6.1.1.11]
Length=585
Score = 35.4 bits (80), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 41/92 (44%), Gaps = 7/92 (7%)
Query 93 VQQLLRRSVAAAPRKWRQLAAGVRGVSEETTTGVLRLLRR-----AAANLLLFPAMNVND 147
V LLR + A K R+L + + V +E T L+L N +L PA N+
Sbjct 221 VNSLLRSVIIGARNKIRELKSELNTVEQELTALRLKLPNLLHPEVTDNNTVLKPAKYSNN 280
Query 148 CVTKSKFDNVYGCRHSLPDAIMRATDVMLGGK 179
V KSK D+V P I RA V + GK
Sbjct 281 AVLKSKLDHVDVINRLCPRYIERA--VSISGK 310
> dre:100150965 retinol dehydrogenase 12-like
Length=296
Score = 32.3 bits (72), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query 171 ATDVMLGGKLAVVCGYGD-VGKGCAAALRGQGCRVIISEIDPICALQAAME 220
++DV L GK A+V G +GK A L +G RVI++ D + A QAA +
Sbjct 13 SSDVRLDGKTAIVTGANTGIGKETAKDLANRGARVILACRDLVKAEQAASD 63
> dre:561703 rictora, si:ch211-14b14.3; rapamycin-insensitive
companion of mTOR a
Length=989
Score = 31.6 bits (70), Expect = 2.5, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 36/75 (48%), Gaps = 17/75 (22%)
Query 141 PAMNVNDCVTKSKFDNVYGCRHSLPDAIMR--------ATDVMLGGKLAVVCGYGDVGKG 192
P+ N + VT+SKF V GC DA + +T+++LGGK + G
Sbjct 843 PSHNHLEVVTQSKFSGVSGCS----DAAVSQGSACSTPSTEIILGGK-----AISEDGPA 893
Query 193 CAAALRGQGCRVIIS 207
C LR + R++I+
Sbjct 894 CRVLLRKEVLRLVIN 908
> cel:F09E10.3 dhs-25; DeHydrogenases, Short chain family member
(dhs-25)
Length=248
Score = 30.8 bits (68), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Query 175 MLGGKLAVVCGYGD-VGKGCAAALRGQGCRVIISEID 210
+LGG++AVV G +GK + L G RV+++++D
Sbjct 4 LLGGRIAVVTGGASGIGKAISQTLAKHGARVVVADLD 40
Lambda K H
0.320 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8274230796
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40