bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3839_orf1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
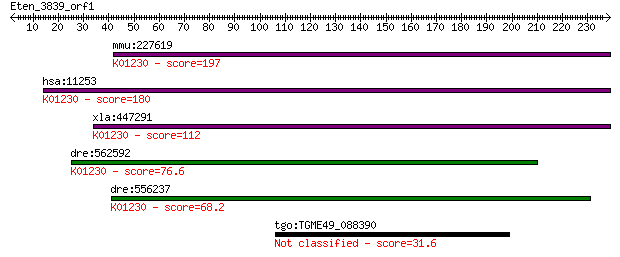
mmu:227619 Man1b1, E430019H13Rik, Gm108, MANA-ER; mannosidase,... 197 3e-50
hsa:11253 MAN1B1, ERMan1, MANA-ER; mannosidase, alpha, class 1... 180 4e-45
xla:447291 man1b1, MGC78858, erman1, mana-er; mannosidase, alp... 112 1e-24
dre:562592 alpha 1,2-mannosidase-like; K01230 mannosyl-oligosa... 76.6 7e-14
dre:556237 man1b1, si:dkey-61l1.3; mannosidase, alpha, class 1... 68.2 2e-11
tgo:TGME49_088390 glycosyl transferase, group 2 family domain-... 31.6 2.8
> mmu:227619 Man1b1, E430019H13Rik, Gm108, MANA-ER; mannosidase,
alpha, class 1B, member 1 (EC:3.2.1.113); K01230 mannosyl-oligosaccharide
alpha-1,2-mannosidase [EC:3.2.1.113]
Length=658
Score = 197 bits (500), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 101/197 (51%), Positives = 127/197 (64%), Gaps = 0/197 (0%)
Query 42 RRDFISVTLSPGRSYDGGQGLRRRSCWRKWKQLSRLQRTVVLFLLVVVMLFGLLSYVHVA 101
RDFISVTLS G SYD + RRRSCWRKWKQLSRLQR V+LF+L ++L G L +H A
Sbjct 12 HRDFISVTLSLGESYDNSKSRRRRSCWRKWKQLSRLQRNVILFVLGFLILCGFLYSLHTA 71
Query 102 DEWTAVDSRSAAAGQMRPANPPVLPAPQKAAENPEAVAGLSPQKPQRHFRRGPPNLQIRA 161
D+W A+ R A +M+ PVLPAPQK + E A + QK QRHFRRGPP+LQIR
Sbjct 72 DQWKALSGRPAEVEKMKQEVLPVLPAPQKESAEQEGFADILSQKRQRHFRRGPPHLQIRP 131
Query 162 PDGDPQERRQDRAQRRAEVVGEAGWGAEAQRDGLSWRGAGTKPQQGTEPPGKKAELPARP 221
P+ ++ QD A+ R +G+A QR +SWRGA +P+Q TE P K+AE +P
Sbjct 132 PNTVSKDGMQDDAKEREAALGKAQQEENTQRTVISWRGAVIEPEQATELPYKRAEASIKP 191
Query 222 SPKAARIQANPAPQNER 238
A++I PAP NER
Sbjct 192 LVLASKIWKEPAPPNER 208
> hsa:11253 MAN1B1, ERMan1, MANA-ER; mannosidase, alpha, class
1B, member 1 (EC:3.2.1.113); K01230 mannosyl-oligosaccharide
alpha-1,2-mannosidase [EC:3.2.1.113]
Length=699
Score = 180 bits (456), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 118/235 (50%), Positives = 148/235 (62%), Gaps = 10/235 (4%)
Query 14 SKQGFLTPPEGEA--LRAPAAVMYPPPAPA-RRDFISVTLSPGRSYDGGQGLRRRSCWRK 70
S+ FLTPP G A A VMYPPP P RDFISVTLS G +YD + RRRSCWRK
Sbjct 15 SQSDFLTPPVGGAPWAVATTVVMYPPPPPPPHRDFISVTLSFGENYDNSKSWRRRSCWRK 74
Query 71 WKQLSRLQRTVVLFLLVVVMLFGLLSYVHVADEWTAVDSRSAAAGQMRP-------ANPP 123
WKQLSRLQR ++LFLL ++ GLL Y+++AD W A+ R +MRP ANPP
Sbjct 75 WKQLSRLQRNMILFLLAFLLFCGLLFYINLADHWKALAFRLEEEQKMRPEIAGLKPANPP 134
Query 124 VLPAPQKAAENPEAVAGLSPQKPQRHFRRGPPNLQIRAPDGDPQERRQDRAQRRAEVVGE 183
VLPAPQKA +PE + +S QK QRH +RGPP+LQIR P D ++ Q+ A +R E +
Sbjct 135 VLPAPQKADTDPENLPEISSQKTQRHIQRGPPHLQIRPPSQDLKDGTQEEATKRQEAPVD 194
Query 184 AGWGAEAQRDGLSWRGAGTKPQQGTEPPGKKAELPARPSPKAARIQANPAPQNER 238
+ QR +SWRGA +P+QGTE P ++AE+P +P AR Q P N R
Sbjct 195 PRPEGDPQRTVISWRGAVIEPEQGTELPSRRAEVPTKPPLPPARTQGTPVHLNYR 249
> xla:447291 man1b1, MGC78858, erman1, mana-er; mannosidase, alpha,
class 1B, member 1 (EC:3.2.1.113); K01230 mannosyl-oligosaccharide
alpha-1,2-mannosidase [EC:3.2.1.113]
Length=641
Score = 112 bits (281), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 78/207 (37%), Positives = 114/207 (55%), Gaps = 18/207 (8%)
Query 34 MYPPPAPARRDFISVTLSPGRSYDGGQGLRRRSCWRKWKQLSRLQRTVVLFLLVVVMLFG 93
MY +P RRDF+SVT+S Y+ +G RR+SCWRKWKQLSR QR ++LFL VV+ +
Sbjct 1 MY---SPQRRDFLSVTVSQAEGYNNSKGWRRKSCWRKWKQLSRFQRNIILFLCVVLAVCT 57
Query 94 LLSYVHVADEWTAVDSRS--AAAGQMRPANPPVLPAPQKAAENPEAVAGLSPQKPQRHFR 151
++S+ ++ + W A+ S+S Q P VLPAP++ ++ + QKP+R +
Sbjct 58 VVSFSNLGEHWRALISQSEEHYESQDGPGVKAVLPAPKREEDSDLMLLKPPAQKPRRPHK 117
Query 152 RGPPNLQIRAPDGDPQERRQDRAQRRAEVVGEAGWGAEAQRDGLSWRGAGTKPQQGTEPP 211
RGPP LQ G ++ +D QR +V E Q+ +SWRGA P Q +EP
Sbjct 118 RGPPILQPPPTPG---QKVEDNEQRPDQV--------ENQQPVISWRGAVIDPVQPSEPT 166
Query 212 GKKAELPARPSPKAARIQANPAPQNER 238
K + +P A + P P +ER
Sbjct 167 VNKVQKATAAAPVAN--EKEPVPISER 191
> dre:562592 alpha 1,2-mannosidase-like; K01230 mannosyl-oligosaccharide
alpha-1,2-mannosidase [EC:3.2.1.113]
Length=682
Score = 76.6 bits (187), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 67/208 (32%), Positives = 101/208 (48%), Gaps = 32/208 (15%)
Query 25 EALRAPAAVMYPPPAPARRDFISVTL--SPGRSYDGGQGLRRRSCWRKWKQLSRLQRTVV 82
E+L MY +P ++DF+S+T+ R Y G+ RR+SCWRKWKQLSRLQR+++
Sbjct 3 ESLSGGLVEMY---SPQKKDFVSLTVREQHNRGYTNGKH-RRQSCWRKWKQLSRLQRSLI 58
Query 83 LFLLVVVMLFGLLSYVHVADEWTAVDSRSAAAGQ-------MRPANPPVLPAPQKAAENP 135
LFLL + + G+LSY + ++W + RS +RP P ++P
Sbjct 59 LFLLAFLFICGILSYSSLTEQWRGISDRSLKEDWDEVDNRGLRPIPPGLIPKLSAGVGVQ 118
Query 136 EAVAGLSP--QKPQRHF------------RRGPPNLQIRAPDGDPQERRQDRAQRRAEVV 181
+ + P QKP +RGPP LQ R D Q + + + +
Sbjct 119 DVQPDVMPELQKPSIPLLPKPPAMRKPPSKRGPPALQKRMNVSDWQLKDGEEGLIKTDED 178
Query 182 GEAGWGAEAQRDGLSWRGAGTKPQQGTE 209
GA+ + +SWRGA + +QGTE
Sbjct 179 -----GAKEAKTVISWRGAMIEAEQGTE 201
> dre:556237 man1b1, si:dkey-61l1.3; mannosidase, alpha, class
1B, member 1 (EC:3.2.1.113); K01230 mannosyl-oligosaccharide
alpha-1,2-mannosidase [EC:3.2.1.113]
Length=632
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 56/196 (28%), Positives = 95/196 (48%), Gaps = 21/196 (10%)
Query 41 ARRDFISVTLSPGRSYDGGQGLRRRSCWRKWKQLSRLQRTVVLFLLVVVMLFGLLSYVHV 100
R+DF+S+TLS SY + RR+SCWRKWKQLSRLQR+++L L+++++ G+ + +
Sbjct 5 TRKDFVSLTLSESGSYSNSKQWRRQSCWRKWKQLSRLQRSLILLTLMLLLVCGIATSPTL 64
Query 101 ADEWTAV---DSRSAAAGQMRPANPPVLPAPQKAAENPEAVAGLSPQKPQRHFRRGPPNL 157
W + D +++ ++ +LP P P + + RRG L
Sbjct 65 IGHWRGMNGDDVKNSFVVDLKNEYRSILPEP--------------PSERKDFDRRGAFGL 110
Query 158 QIRAPDGDPQERRQDRAQRRAEVVGEAGWGAEAQRDGLSWRGAGTKPQQGTEPPGKKAEL 217
+ + G Q + A G+E Q+ +SWRGA + +Q ++ K E
Sbjct 111 E-KLLQGKVNVSGVLGDQNHGLDLKNADKGSEGQKPIISWRGAVIEEEQASDTDTKDTEN 169
Query 218 PARPSPKA---ARIQA 230
P+ A +R++A
Sbjct 170 QGDPASLALAESRLEA 185
> tgo:TGME49_088390 glycosyl transferase, group 2 family domain-containing
protein (EC:2.1.1.43)
Length=752
Score = 31.6 bits (70), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 47/104 (45%), Gaps = 14/104 (13%)
Query 106 AVDSRSAAAGQMRPANPPVLPAPQKAAENPEAVAGLSPQKPQRHF-------RRGPPNLQ 158
AV+S A +R +N P +A E P A++ LSP H RRG +
Sbjct 98 AVNSLLAQTFVVRTSNLPASGRSAEAGEKPPALSALSPSAGTAHASEQKKEGRRGDGKPR 157
Query 159 IRAPDGDPQERRQDRAQRR---AEVVGEAGWGA-EAQRDGLSWR 198
+GD +E +QDRA+ R E GE G E + +G WR
Sbjct 158 ---KEGDSKEEKQDRAEDRDAKEERQGEEDEGYKEREEEGDMWR 198
Lambda K H
0.315 0.132 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8274230796
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40