bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3832_orf4
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
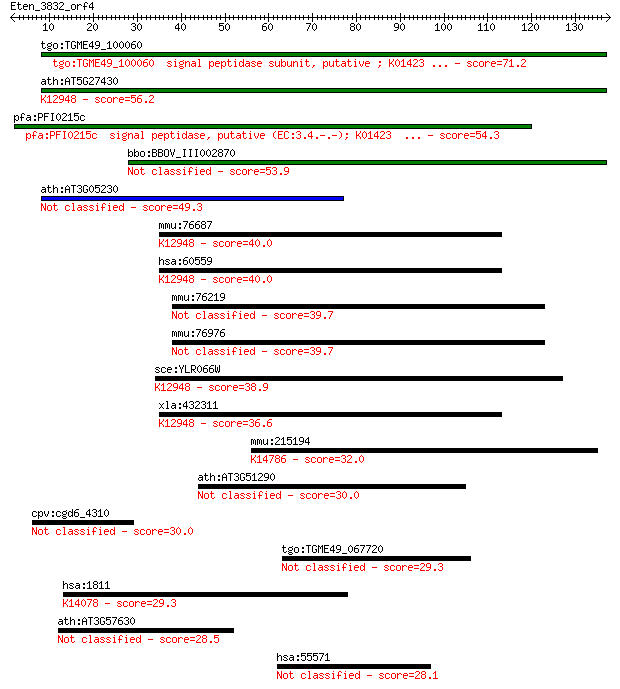
tgo:TGME49_100060 signal peptidase subunit, putative ; K01423 ... 71.2 1e-12
ath:AT5G27430 signal peptidase subunit family protein; K12948 ... 56.2 3e-08
pfa:PFI0215c signal peptidase, putative (EC:3.4.-.-); K01423 ... 54.3 1e-07
bbo:BBOV_III002870 17.m07272; signal peptidase family protein 53.9 2e-07
ath:AT3G05230 signal peptidase subunit family protein 49.3 3e-06
mmu:76687 Spcs3, 1810011E08Rik; signal peptidase complex subun... 40.0 0.002
hsa:60559 SPCS3, DKFZp564J1864, FLJ22649, PRO3567, SPC22/23, S... 40.0 0.002
mmu:76219 Arxes1, 6530401D17Rik, Spcs3; adipocyte-related X-ch... 39.7 0.003
mmu:76976 Arxes2, 2900062L11Rik, Spcs3; adipocyte-related X-ch... 39.7 0.003
sce:YLR066W SPC3; Spc3p; K12948 signal peptidase complex subun... 38.9 0.005
xla:432311 spcs3, MGC79052; signal peptidase complex subunit 3... 36.6 0.024
mmu:215194 Kri1, MGC29361; KRI1 homolog (S. cerevisiae); K1478... 32.0 0.61
ath:AT3G51290 proline-rich family protein 30.0 2.1
cpv:cgd6_4310 hypothetical protein 30.0 2.2
tgo:TGME49_067720 hypothetical protein 29.3 3.9
hsa:1811 SLC26A3, CLD, DRA; solute carrier family 26, member 3... 29.3 4.1
ath:AT3G57630 exostosin family protein 28.5 6.7
hsa:55571 C2orf29; chromosome 2 open reading frame 29 28.1
> tgo:TGME49_100060 signal peptidase subunit, putative ; K01423
[EC:3.4.-.-]
Length=175
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 41/129 (31%), Positives = 63/129 (48%), Gaps = 0/129 (0%)
Query 8 SGSIPVKDVYGFSYNSPLNGDQAVFFLDKKADLGGLFQGNAKQGSLFVGAEYETPQPPTN 67
+G + + +VY F N+ L G+QA L+ +ADL F N +Q ++V YETP+ P N
Sbjct 38 TGKVSIAEVYEFGVNNALQGEQAQVALNIQADLTSCFNWNTQQLFVYVIVRYETPKNPRN 97
Query 68 QGGGFAPIIMDDGVAVIDGPNPPGKSHFRNKGGGLGGPEVTVNLQVFSPPIGGRMYPQTF 127
+ + II D A+ID K R+ G L VTV L+ P+ G +
Sbjct 98 EVIVWDRIITDPDDAIIDFEGVINKYPLRDNGRSLRNRTVTVALEYAYHPVVGVIKSGHV 157
Query 128 ARNPFRMPG 136
A + + +P
Sbjct 158 ASSTYTLPS 166
> ath:AT5G27430 signal peptidase subunit family protein; K12948
signal peptidase complex subunit 3 [EC:3.4.-.-]
Length=167
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 54/129 (41%), Gaps = 2/129 (1%)
Query 8 SGSIPVKDVYGFSYNSPLNGDQAVFFLDKKADLGGLFQGNAKQGSLFVGAEYETPQPPTN 67
S I + ++ F P D+ L+ ADL LF N KQ FV AEYET + N
Sbjct 38 SAQIQILNINWFQ-KQPHGNDEVSLTLNITADLQSLFTWNTKQVFAFVAAEYETSKNALN 96
Query 68 QGGGFAPIIMDDGVAVIDGPNPPGKSHFRNKGGGLGGPEVTVNLQVFSPPIGGRMYPQTF 127
Q + II + A K F ++G L G + + L P G+M+
Sbjct 97 QVSLWDAIIPEKEHAKF-WIQISNKYRFIDQGHNLRGKDFNLTLHWHVMPKTGKMFADKI 155
Query 128 ARNPFRMPG 136
+ +R+P
Sbjct 156 VMSGYRLPN 164
> pfa:PFI0215c signal peptidase, putative (EC:3.4.-.-); K01423
[EC:3.4.-.-]
Length=185
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 36/121 (29%), Positives = 54/121 (44%), Gaps = 5/121 (4%)
Query 2 FPRGPISGSIPVKDVYGFSYNSPLNGDQAVFFLDKKADLGGLFQGNAKQGSLFVGAEYET 61
F I +I VK + F YN +N D+AV LD D+ F N KQ ++V YET
Sbjct 34 FDEKEIKTNIQVKSIKRFVYNRYINADEAVLSLDVSYDMRKAFNWNLKQLFVYVLVTYET 93
Query 62 PQPPTNQGGGFAPIIMDDGVAVIDGPNPPGKSHFRNKGGGLGGPEVTVNLQV---FSPPI 118
P+ N+ I+ + A + N K ++ GL ++LQV + P +
Sbjct 94 PKKIKNEVIIQDYIVKNKKQAKKNYRNFITKYSLKDYYNGLRNN--LIHLQVCYKYMPIV 151
Query 119 G 119
G
Sbjct 152 G 152
> bbo:BBOV_III002870 17.m07272; signal peptidase family protein
Length=171
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 46/109 (42%), Gaps = 0/109 (0%)
Query 28 DQAVFFLDKKADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGFAPIIMDDGVAVIDGP 87
D+A F L+ DL +F +A L+ YETP+ P N+ F II A G
Sbjct 59 DRAAFELNLSYDLRDVFDWSANVIFLYATVNYETPKHPVNELIIFDKIITSKEEAYEPGA 118
Query 88 NPPGKSHFRNKGGGLGGPEVTVNLQVFSPPIGGRMYPQTFARNPFRMPG 136
+ K + + L VT+ L PIGG + A + F MP
Sbjct 119 DIVSKYYMIDYARSLRKARVTLRLHYCFVPIGGLIKSYQLAESVFTMPS 167
> ath:AT3G05230 signal peptidase subunit family protein
Length=136
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Query 8 SGSIPVKDVYGFSYNSPLNGDQAVFFLDKKADLGGLFQGNAKQGSLFVGAEYETPQPPTN 67
S I + ++ F S N D+ LD ADL LF N KQ +FV AEYETP+ N
Sbjct 38 SAEIQILNINRFKKQSHGN-DEVSLTLDISADLQSLFTWNTKQVFVFVAAEYETPKNSLN 96
Query 68 QGGGFAPII 76
Q + II
Sbjct 97 QVSLWDAII 105
> mmu:76687 Spcs3, 1810011E08Rik; signal peptidase complex subunit
3 homolog (S. cerevisiae); K12948 signal peptidase complex
subunit 3 [EC:3.4.-.-]
Length=180
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 39/86 (45%), Gaps = 15/86 (17%)
Query 35 DKKADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGFAPIIMDDGVAVIDGPNP----- 89
D ADL +F N KQ L++ AEY T NQ +++ D + V+ G NP
Sbjct 69 DITADLENIFDWNVKQLFLYLSAEYSTKNNALNQ------VVLWDKI-VLRGDNPKLLLK 121
Query 90 --PGKSHFRNKGGGL-GGPEVTVNLQ 112
K F + G GL G VT+ L
Sbjct 122 DMKTKYFFFDDGNGLKGNRNVTLTLS 147
> hsa:60559 SPCS3, DKFZp564J1864, FLJ22649, PRO3567, SPC22/23,
SPC3, YLR066W; signal peptidase complex subunit 3 homolog (S.
cerevisiae); K12948 signal peptidase complex subunit 3 [EC:3.4.-.-]
Length=180
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 39/86 (45%), Gaps = 15/86 (17%)
Query 35 DKKADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGFAPIIMDDGVAVIDGPNP----- 89
D ADL +F N KQ L++ AEY T NQ +++ D + V+ G NP
Sbjct 69 DITADLENIFDWNVKQLFLYLSAEYSTKNNALNQ------VVLWDKI-VLRGDNPKLLLK 121
Query 90 --PGKSHFRNKGGGL-GGPEVTVNLQ 112
K F + G GL G VT+ L
Sbjct 122 DMKTKYFFFDDGNGLKGNRNVTLTLS 147
> mmu:76219 Arxes1, 6530401D17Rik, Spcs3; adipocyte-related X-chromosome
expressed sequence 1
Length=180
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 38/86 (44%), Gaps = 1/86 (1%)
Query 38 ADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGFAPIIMDDGVAVIDGPNPPGKSHFRN 97
ADL F N KQ L++ AEY T NQ + I++ ++ + K F +
Sbjct 72 ADLEKTFDWNVKQLFLYLSAEYSTKSNAVNQVVLWDKILLRGENPKLNLKDVKSKYFFFD 131
Query 98 KGGGL-GGPEVTVNLQVFSPPIGGRM 122
G GL G VT+ L PI G +
Sbjct 132 DGHGLKGNRNVTLTLSWQVIPIAGIL 157
> mmu:76976 Arxes2, 2900062L11Rik, Spcs3; adipocyte-related X-chromosome
expressed sequence 2
Length=180
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 38/86 (44%), Gaps = 1/86 (1%)
Query 38 ADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGFAPIIMDDGVAVIDGPNPPGKSHFRN 97
ADL F N KQ L++ AEY T NQ + I++ ++ + K F +
Sbjct 72 ADLEKTFDWNVKQLFLYLSAEYSTKSNAVNQVVLWDKILLRGENPKLNLKDVKSKYFFFD 131
Query 98 KGGGL-GGPEVTVNLQVFSPPIGGRM 122
G GL G VT+ L PI G +
Sbjct 132 DGHGLKGNRNVTLTLSWQVIPIAGIL 157
> sce:YLR066W SPC3; Spc3p; K12948 signal peptidase complex subunit
3 [EC:3.4.-.-]
Length=184
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 42/95 (44%), Gaps = 2/95 (2%)
Query 34 LDKKADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGFAPIIMDDGVAVIDGPNPPGK- 92
D DL LF N KQ +++ AEY + + T++ + II AVID + K
Sbjct 76 FDLNTDLTPLFNWNTKQVFVYLTAEYNSTEKITSEVTFWDKIIKSKDDAVIDVNDLRSKY 135
Query 93 SHFRNKGGGLGGPEVTVNLQV-FSPPIGGRMYPQT 126
S + + G G ++ L P +G Y +T
Sbjct 136 SIWDIEDGKFEGKDLVFKLHWNVQPWVGLLTYGET 170
> xla:432311 spcs3, MGC79052; signal peptidase complex subunit
3 homolog; K12948 signal peptidase complex subunit 3 [EC:3.4.-.-]
Length=180
Score = 36.6 bits (83), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 34/79 (43%), Gaps = 1/79 (1%)
Query 35 DKKADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGFAPIIMDDGVAVIDGPNPPGKSH 94
D ADL +F N KQ +++ AEY T NQ + II+ + K
Sbjct 69 DINADLQPIFDWNVKQLFIYLSAEYSTRSNTLNQVVLWDKIILRGDNPKLSLKEMKSKYF 128
Query 95 FRNKGGGL-GGPEVTVNLQ 112
F + G GL G +T+ L
Sbjct 129 FFDDGNGLKGNRNITLTLS 147
> mmu:215194 Kri1, MGC29361; KRI1 homolog (S. cerevisiae); K14786
protein KRI1
Length=705
Score = 32.0 bits (71), Expect = 0.61, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 34/79 (43%), Gaps = 12/79 (15%)
Query 56 GAEYETPQPPTNQGGGFAPIIMDDGVAVIDGPNPPGKSHFRNKGGGLGGPEVTVNLQVFS 115
GA+ +TPQ T + ++G+A P P ++K L GP VT+ FS
Sbjct 623 GADQKTPQAET-------LVSKEEGLAHTSCPEKPASQKQKSKKARLLGPTVTLGGHKFS 675
Query 116 PPIGGRMYPQTFARNPFRM 134
R Q F NP R+
Sbjct 676 -----RQRLQAFGLNPKRL 689
> ath:AT3G51290 proline-rich family protein
Length=602
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 28/62 (45%), Gaps = 16/62 (25%)
Query 44 FQGNAKQGSLFVGAEYETPQPPTN-QGGGFAPIIMDDGVAVIDGPNPPGKSHFRNKGGGL 102
F G++K G ++ + YE PT+ GFAP + S +RN GG +
Sbjct 236 FSGHSKSGKMYSSSNYECNLNPTSFWTRGFAPSKL---------------SEYRNAGGVI 280
Query 103 GG 104
GG
Sbjct 281 GG 282
> cpv:cgd6_4310 hypothetical protein
Length=1931
Score = 30.0 bits (66), Expect = 2.2, Method: Composition-based stats.
Identities = 13/23 (56%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 6 PISGSIPVKDVYGFSYNSPLNGD 28
P+S SI +KD+ S +SPLNGD
Sbjct 1594 PLSISINIKDIDNLSDHSPLNGD 1616
> tgo:TGME49_067720 hypothetical protein
Length=1290
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 2/43 (4%)
Query 63 QPPTNQGGGFAPIIMDDGVAVIDGPNPPGKSHFRNKGGGLGGP 105
QPPT G AP+ A + GP+PP ++ N GGP
Sbjct 347 QPPTGTGANRAPLRAGLARAGLQGPSPPAQNGAANSRS--GGP 387
> hsa:1811 SLC26A3, CLD, DRA; solute carrier family 26, member
3; K14078 solute carrier family 26 (chloride anion exchanger),
member 3
Length=764
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 30/65 (46%), Gaps = 0/65 (0%)
Query 13 VKDVYGFSYNSPLNGDQAVFFLDKKADLGGLFQGNAKQGSLFVGAEYETPQPPTNQGGGF 72
V VY Y+ PL+G+Q + L + G+F+G A +L A E+ T G
Sbjct 357 VASVYSLKYDYPLDGNQELIALGLGNIVCGVFRGFAGSTALSRSAVQESTGGKTQIAGLI 416
Query 73 APIIM 77
II+
Sbjct 417 GAIIV 421
> ath:AT3G57630 exostosin family protein
Length=793
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 12 PVKDVYGFSYNSPLNGDQAVFFLDKKADLGGLFQGNAKQG 51
PV + GF NSP N D+ K DL L ++KQG
Sbjct 198 PVPESCGFQINSPTNPDEPKMTDWSKPDLDILTTNSSKQG 237
> hsa:55571 C2orf29; chromosome 2 open reading frame 29
Length=510
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 62 PQPPTNQGGGFAPIIMDDGVAVIDGPNPPGKSHFR 96
P P ++ G + + A++ GP PP +SHFR
Sbjct 247 PDPDSSNSGFDSSVASQITEALVSGPKPPIESHFR 281
Lambda K H
0.317 0.144 0.450
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40