bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3826_orf4
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
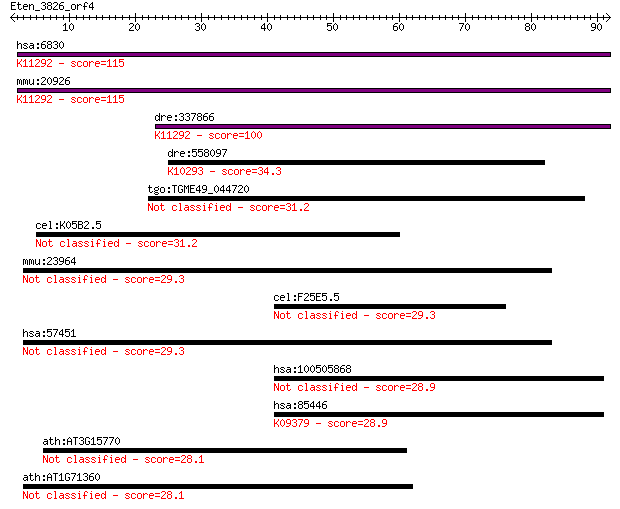
hsa:6830 SUPT6H, KIAA0162, MGC87943, SPT6, SPT6H, emb-5; suppr... 115 4e-26
mmu:20926 Supt6h, 5131400N11Rik, AI132449, SPT6, mKIAA0162; su... 115 5e-26
dre:337866 supt6h, Spt6, fj42h11, wu:fj42h11; suppressor of Ty... 100 9e-22
dre:558097 fbxo7, MGC114096, si:ch211-286m4.2, zgc:114096; F-b... 34.3 0.11
tgo:TGME49_044720 hypothetical protein 31.2 0.90
cel:K05B2.5 pes-22; Patterned Expression Site family member (p... 31.2 0.93
mmu:23964 Odz2, 2610040L17Rik, 9330187F13Rik, D3Bwg1534e, Odz3... 29.3 2.8
cel:F25E5.5 hypothetical protein 29.3 2.9
hsa:57451 ODZ2, DKFZp686A1568, TEN-M2, TNM2; odz, odd Oz/ten-m... 29.3 3.3
hsa:100505868 zinc finger homeobox protein 2-like 28.9 3.9
hsa:85446 ZFHX2, FLJ17566, FLJ42478, KIAA1056, KIAA1762, MGC14... 28.9 4.1
ath:AT3G15770 hypothetical protein 28.1 6.6
ath:AT1G71360 hypothetical protein 28.1 7.4
> hsa:6830 SUPT6H, KIAA0162, MGC87943, SPT6, SPT6H, emb-5; suppressor
of Ty 6 homolog (S. cerevisiae); K11292 transcription
elongation factor SPT6
Length=1726
Score = 115 bits (288), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 71/90 (78%), Positives = 79/90 (87%), Gaps = 0/90 (0%)
Query 2 QLQASTPPQFVQAQAQPSSSSRQRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQRM 61
QLQAST PQ QAQ QPSSSSRQRQ+QPKS++H+ I+WGKMAEQWLQ+KEAERRKQKQR+
Sbjct 1637 QLQASTTPQSAQAQPQPSSSSRQRQQQPKSNSHAAIDWGKMAEQWLQEKEAERRKQKQRL 1696
Query 62 TPWPSPSPMIESPPKFIAGDSPPLLGEMDR 91
TP PSPSPMIES P IAGD+ PLL EMDR
Sbjct 1697 TPRPSPSPMIESTPMSIAGDATPLLDEMDR 1726
> mmu:20926 Supt6h, 5131400N11Rik, AI132449, SPT6, mKIAA0162;
suppressor of Ty 6 homolog (S. cerevisiae); K11292 transcription
elongation factor SPT6
Length=1726
Score = 115 bits (287), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 71/90 (78%), Positives = 79/90 (87%), Gaps = 0/90 (0%)
Query 2 QLQASTPPQFVQAQAQPSSSSRQRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQRM 61
QLQAST PQ QAQ QPSSSSRQRQ+QPKS++H+ I+WGKMAEQWLQ+KEAERRKQKQR+
Sbjct 1637 QLQASTTPQSTQAQPQPSSSSRQRQQQPKSNSHAAIDWGKMAEQWLQEKEAERRKQKQRL 1696
Query 62 TPWPSPSPMIESPPKFIAGDSPPLLGEMDR 91
TP PSPSPMIES P IAGD+ PLL EMDR
Sbjct 1697 TPRPSPSPMIESTPMSIAGDATPLLDEMDR 1726
> dre:337866 supt6h, Spt6, fj42h11, wu:fj42h11; suppressor of
Ty 6 homolog; K11292 transcription elongation factor SPT6
Length=1726
Score = 100 bits (250), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 50/70 (71%), Positives = 57/70 (81%), Gaps = 1/70 (1%)
Query 23 RQRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQ-RMTPWPSPSPMIESPPKFIAGD 81
R R QPK+ +H+ ++WGKMAEQWLQ+KEAERRKQK RMTP PSPSPMIES P IAGD
Sbjct 1657 RVRTPQPKASSHTAVDWGKMAEQWLQEKEAERRKQKTPRMTPRPSPSPMIESTPMSIAGD 1716
Query 82 SPPLLGEMDR 91
+ PLL EMDR
Sbjct 1717 ATPLLDEMDR 1726
> dre:558097 fbxo7, MGC114096, si:ch211-286m4.2, zgc:114096; F-box
protein 7; K10293 F-box protein 7
Length=486
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 9/66 (13%)
Query 25 RQEQPKSHNHSTIEWGKMAEQ-WLQKKEAERRKQK-------QRMTPWP-SPSPMIESPP 75
R P H H +W ++ +Q + Q+KEA RR + + P+P SP+P+ PP
Sbjct 375 RVSFPAGHQHRDTDWRELYKQKYRQRKEAARRGRHWFYPPPISPLIPFPSSPAPLPLYPP 434
Query 76 KFIAGD 81
I GD
Sbjct 435 GIIGGD 440
> tgo:TGME49_044720 hypothetical protein
Length=2522
Score = 31.2 bits (69), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 35/68 (51%), Gaps = 16/68 (23%)
Query 22 SRQRQEQPKSHNHSTIEWGKMAE--QWLQKKEAERRKQKQRMTPWPSPSPMIESPPKFIA 79
S Q +E+PK+H S + W + AE Q ++ E+R+ +E+ + +A
Sbjct 1825 SLQDEERPKTHRGSDVTWEQQAELIQACRRLALEQRE--------------LEAKIQLLA 1870
Query 80 GDSPPLLG 87
G SPP+LG
Sbjct 1871 GGSPPILG 1878
> cel:K05B2.5 pes-22; Patterned Expression Site family member
(pes-22)
Length=777
Score = 31.2 bits (69), Expect = 0.93, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Query 5 ASTPPQFVQAQAQPSSSSR-----QRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQ 59
A T Q Q Q + S + +R E+ + TI+ K Q LQ E E+RKQKQ
Sbjct 696 ADTADGMPQLQDQDNVSGKLSVLTERSEEALTDYQRTIQSMKQQHQLLQDLEEEKRKQKQ 755
> mmu:23964 Odz2, 2610040L17Rik, 9330187F13Rik, D3Bwg1534e, Odz3,
Ten-m2, mKIAA1127; odd Oz/ten-m homolog 2 (Drosophila)
Length=2764
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 32/82 (39%), Gaps = 4/82 (4%)
Query 3 LQASTPPQFVQAQAQPS--SSSRQRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQR 60
L+A + PQ + P+ S S R P HNH+ A + RR Q
Sbjct 225 LRACSGPQQASSSGPPNHHSQSTLRPPLPPPHNHTLSHHHSSANSLNRNSLTNRRSQIH- 283
Query 61 MTPWPSPSPMIESPPKFIAGDS 82
P P+P+ + +P DS
Sbjct 284 -APAPAPNDLATTPESVQLQDS 304
> cel:F25E5.5 hypothetical protein
Length=409
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 11/35 (31%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 41 KMAEQWLQKKEAERRKQKQRMTPWPSPSPMIESPP 75
+ AE WL++ E ++ + M+ +P+ + IES P
Sbjct 173 RFAESWLKRSRFEVKEIRVEMSKYPTDTSQIESLP 207
> hsa:57451 ODZ2, DKFZp686A1568, TEN-M2, TNM2; odz, odd Oz/ten-m
homolog 2 (Drosophila)
Length=2765
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 32/82 (39%), Gaps = 4/82 (4%)
Query 3 LQASTPPQFVQAQAQPS--SSSRQRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQR 60
L+A + PQ + P+ S S R P HNH+ A + RR Q
Sbjct 225 LRACSGPQQASSSGPPNHHSQSTLRPPLPPPHNHTLSHHHSSANSLNRNSLTNRRSQIH- 283
Query 61 MTPWPSPSPMIESPPKFIAGDS 82
P P+P+ + +P DS
Sbjct 284 -APAPAPNDLATTPESVQLQDS 304
> hsa:100505868 zinc finger homeobox protein 2-like
Length=2706
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 27/58 (46%), Gaps = 13/58 (22%)
Query 41 KMAEQWLQKKEAERRKQKQRMTPWPSPSPMIESP--------PKFIAGDSPPLLGEMD 90
++ + W Q A RK + R TP PSP ++ P PKF LLG++D
Sbjct 2033 RVVQVWFQNTRARERKGQFRSTPGGVPSPAVKPPATATPASLPKFNL-----LLGKVD 2085
> hsa:85446 ZFHX2, FLJ17566, FLJ42478, KIAA1056, KIAA1762, MGC141820,
ZFH-5, ZNF409; zinc finger homeobox 2; K09379 zinc finger
homeobox protein 2
Length=2572
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 27/58 (46%), Gaps = 13/58 (22%)
Query 41 KMAEQWLQKKEAERRKQKQRMTPWPSPSPMIESP--------PKFIAGDSPPLLGEMD 90
++ + W Q A RK + R TP PSP ++ P PKF LLG++D
Sbjct 1899 RVVQVWFQNTRARERKGQFRSTPGGVPSPAVKPPATATPASLPKFNL-----LLGKVD 1951
> ath:AT3G15770 hypothetical protein
Length=161
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 4/55 (7%)
Query 6 STPPQFVQAQAQPSSSSRQRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQR 60
ST + +Q SSS+ E + NH + W + +QW+ K +E RK R
Sbjct 67 STTNLTIDSQGCGSSSN----EPAEFVNHGLVLWNQTRQQWVGDKRSESRKSVGR 117
> ath:AT1G71360 hypothetical protein
Length=596
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 27/59 (45%), Gaps = 0/59 (0%)
Query 3 LQASTPPQFVQAQAQPSSSSRQRQEQPKSHNHSTIEWGKMAEQWLQKKEAERRKQKQRM 61
L+AS + V+ ++R+E K +W E L+K E E+ K K+R+
Sbjct 473 LEASKREKEVETMRLEVEGMKEREENTKKEAMEMRKWRMRVETELEKAENEKEKVKERL 531
Lambda K H
0.308 0.123 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007827920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40