bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3797_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
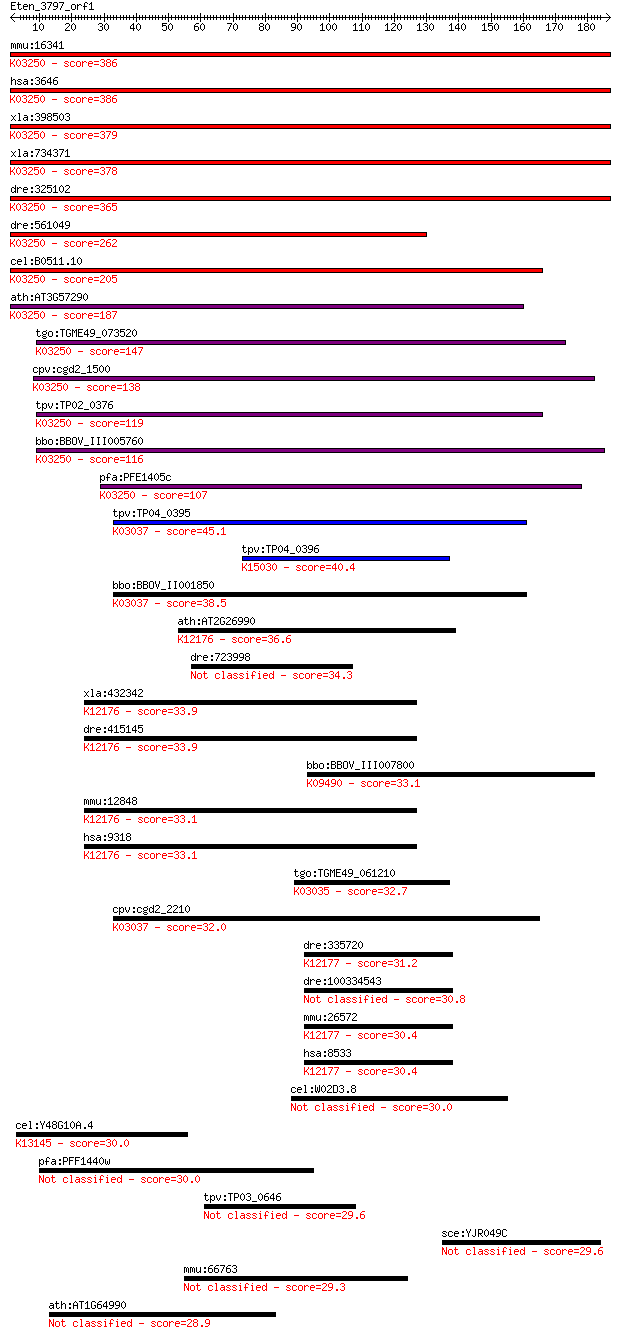
mmu:16341 Eif3e, 48kDa, Eif3s6, Int6, eIF3-p46, eIF3-p48; euka... 386 3e-107
hsa:3646 EIF3E, EIF3-P48, EIF3S6, INT6, eIF3-p46; eukaryotic t... 386 3e-107
xla:398503 eif3e-a, eif3s6-A; eukaryotic translation initiatio... 379 4e-105
xla:734371 eif3e-b, MGC84920, eif3s6-B; eukaryotic translation... 378 7e-105
dre:325102 eif3ea, MGC63821, eIF3-p48, eif3s6, eif3s6a, fc55e0... 365 4e-101
dre:561049 eif3eb, eif3s6b, im:7147439; eukaryotic translation... 262 4e-70
cel:B0511.10 eif-3.E; Eukaryotic Initiation Factor family memb... 205 7e-53
ath:AT3G57290 EIF3E; EIF3E (EUKARYOTIC TRANSLATION INITIATION ... 187 2e-47
tgo:TGME49_073520 proteasome PCI domain-containing protein ; K... 147 2e-35
cpv:cgd2_1500 possible translation initiation factor with poss... 138 9e-33
tpv:TP02_0376 eukaryotic translation initiation factor 3 subun... 119 5e-27
bbo:BBOV_III005760 17.m07513; eukaryotic translation initiatio... 116 4e-26
pfa:PFE1405c eukaryotic translation initiation factor 3, subun... 107 2e-23
tpv:TP04_0395 26S proteasome regulatory subunit; K03037 26S pr... 45.1 1e-04
tpv:TP04_0396 hypothetical protein; K15030 translation initiat... 40.4 0.003
bbo:BBOV_II001850 18.m06141; PCI domain containing 26S proteas... 38.5 0.012
ath:AT2G26990 FUS12; FUS12 (FUSCA 12); K12176 COP9 signalosome... 36.6 0.051
dre:723998 sb:cb336; zgc:136894 34.3 0.23
xla:432342 cops2, csn2; COP9 constitutive photomorphogenic hom... 33.9 0.32
dre:415145 cops2, wu:fc05f05, wu:fc06g10, wu:fc60f04, wu:fl94f... 33.9 0.35
bbo:BBOV_III007800 17.m07684; heat shock protein 70 precursor;... 33.1 0.50
mmu:12848 Cops2, AI315723, C85265, Csn2, Sgn2, Trip15, alien-l... 33.1 0.54
hsa:9318 COPS2, ALIEN, CSN2, SGN2, TRIP15; COP9 constitutive p... 33.1 0.62
tgo:TGME49_061210 26S proteasome non-ATPase regulatory subunit... 32.7 0.74
cpv:cgd2_2210 proteasome regulatory subunit Rpn7/26S proteasom... 32.0 1.2
dre:335720 cops3, wu:fc32a02, wu:fk59f01, zgc:111903, zgc:5564... 31.2 1.9
dre:100334543 COP9 signalosome complex subunit 3-like 30.8 2.5
mmu:26572 Cops3, Csn3, Sgn3; COP9 (constitutive photomorphogen... 30.4 3.2
hsa:8533 COPS3, CSN3, SGN3; COP9 constitutive photomorphogenic... 30.4 3.6
cel:W02D3.8 smg-5; Suppressor with Morphological effect on Gen... 30.0 4.3
cel:Y48G10A.4 hypothetical protein; K13145 integrator complex ... 30.0 5.0
pfa:PFF1440w SET domain protein, putative 30.0 5.0
tpv:TP03_0646 hypothetical protein 29.6 6.7
sce:YJR049C UTR1; ATP-NADH kinase; phosphorylates both NAD and... 29.6 6.9
mmu:66763 4933425L06Rik, MGC141320; RIKEN cDNA 4933425L06 gene 29.3 8.6
ath:AT1G64990 GTG1; GTG1 (GPCR-TYPE G PROTEIN 1) 28.9 9.7
> mmu:16341 Eif3e, 48kDa, Eif3s6, Int6, eIF3-p46, eIF3-p48; eukaryotic
translation initiation factor 3, subunit E; K03250
translation initiation factor 3 subunit E
Length=445
Score = 386 bits (991), Expect = 3e-107, Method: Compositional matrix adjust.
Identities = 185/186 (99%), Positives = 185/186 (99%), Gaps = 0/186 (0%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL
Sbjct 260 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 319
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR
Sbjct 320 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 379
Query 121 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAA 180
LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWA
Sbjct 380 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAT 439
Query 181 QDSGFY 186
QDSGFY
Sbjct 440 QDSGFY 445
> hsa:3646 EIF3E, EIF3-P48, EIF3S6, INT6, eIF3-p46; eukaryotic
translation initiation factor 3, subunit E; K03250 translation
initiation factor 3 subunit E
Length=445
Score = 386 bits (991), Expect = 3e-107, Method: Compositional matrix adjust.
Identities = 185/186 (99%), Positives = 185/186 (99%), Gaps = 0/186 (0%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL
Sbjct 260 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 319
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR
Sbjct 320 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 379
Query 121 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAA 180
LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWA
Sbjct 380 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAT 439
Query 181 QDSGFY 186
QDSGFY
Sbjct 440 QDSGFY 445
> xla:398503 eif3e-a, eif3s6-A; eukaryotic translation initiation
factor 3, subunit E; K03250 translation initiation factor
3 subunit E
Length=446
Score = 379 bits (973), Expect = 4e-105, Method: Compositional matrix adjust.
Identities = 180/186 (96%), Positives = 184/186 (98%), Gaps = 0/186 (0%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL
Sbjct 261 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 320
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
VNDFFLVACLEDFIENARLFIFETFCRIHQCISI+MLADKLNMTPEEAERWIVNLIRNAR
Sbjct 321 VNDFFLVACLEDFIENARLFIFETFCRIHQCISISMLADKLNMTPEEAERWIVNLIRNAR 380
Query 121 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAA 180
LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSL+FRSQMLAMNIEKK+N NSR+EAP WAA
Sbjct 381 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLAFRSQMLAMNIEKKMNTNSRTEAPTWAA 440
Query 181 QDSGFY 186
QDSGFY
Sbjct 441 QDSGFY 446
> xla:734371 eif3e-b, MGC84920, eif3s6-B; eukaryotic translation
initiation factor 3, subunit E; K03250 translation initiation
factor 3 subunit E
Length=446
Score = 378 bits (970), Expect = 7e-105, Method: Compositional matrix adjust.
Identities = 180/186 (96%), Positives = 184/186 (98%), Gaps = 0/186 (0%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYV+FDFDGAQKKLRECESVL
Sbjct 261 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVHFDFDGAQKKLRECESVL 320
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
VNDFFLVACLEDFIENARLFIFETFCRIHQCISI+MLADKLNMTPEEAERWIVNLIRNAR
Sbjct 321 VNDFFLVACLEDFIENARLFIFETFCRIHQCISISMLADKLNMTPEEAERWIVNLIRNAR 380
Query 121 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAA 180
LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSL+FRSQMLAMNIEKK NQNSR+EAP WAA
Sbjct 381 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLAFRSQMLAMNIEKKSNQNSRTEAPTWAA 440
Query 181 QDSGFY 186
QDSGFY
Sbjct 441 QDSGFY 446
> dre:325102 eif3ea, MGC63821, eIF3-p48, eif3s6, eif3s6a, fc55e01,
wu:fc55e01, zgc:63821; eukaryotic translation initiation
factor 3, subunit E, a; K03250 translation initiation factor
3 subunit E
Length=446
Score = 365 bits (937), Expect = 4e-101, Method: Compositional matrix adjust.
Identities = 174/186 (93%), Positives = 180/186 (96%), Gaps = 0/186 (0%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFD AQKKLRECE+VL
Sbjct 261 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDSAQKKLRECEAVL 320
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
VNDFFLVACLEDFIENARLFIFETFCRIHQCISI MLADKLNMTPEEAERWIVNLIRNAR
Sbjct 321 VNDFFLVACLEDFIENARLFIFETFCRIHQCISIGMLADKLNMTPEEAERWIVNLIRNAR 380
Query 121 LDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAA 180
LDAKIDSKLGHVVMGNNA+SPYQQVIEKTKSLSFRSQMLAMNIEKK + +R+E PNWAA
Sbjct 381 LDAKIDSKLGHVVMGNNAISPYQQVIEKTKSLSFRSQMLAMNIEKKQSNANRNETPNWAA 440
Query 181 QDSGFY 186
QD+GFY
Sbjct 441 QDAGFY 446
> dre:561049 eif3eb, eif3s6b, im:7147439; eukaryotic translation
initiation factor 3, subunit E, b; K03250 translation initiation
factor 3 subunit E
Length=401
Score = 262 bits (670), Expect = 4e-70, Method: Compositional matrix adjust.
Identities = 126/129 (97%), Positives = 128/129 (99%), Gaps = 0/129 (0%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFD AQ+KLRECESVL
Sbjct 254 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDSAQRKLRECESVL 313
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
VNDFFLVACLEDFIENARLFIFETFCRIHQCISI+MLADKLNMTPEEAERWIVNLIRNAR
Sbjct 314 VNDFFLVACLEDFIENARLFIFETFCRIHQCISISMLADKLNMTPEEAERWIVNLIRNAR 373
Query 121 LDAKIDSKL 129
LDAKIDSKL
Sbjct 374 LDAKIDSKL 382
> cel:B0511.10 eif-3.E; Eukaryotic Initiation Factor family member
(eif-3.E); K03250 translation initiation factor 3 subunit
E
Length=432
Score = 205 bits (521), Expect = 7e-53, Method: Compositional matrix adjust.
Identities = 100/166 (60%), Positives = 131/166 (78%), Gaps = 3/166 (1%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
AV+T+K +++ LKDLVKVI E ++YKDP+T+F+ CLY+ +DFD AQ+ L++CE VL
Sbjct 260 AVVTSKS--RQKNSLKDLVKVIDIERHSYKDPVTDFLTCLYIKYDFDEAQEMLQKCEEVL 317
Query 61 VNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNAR 120
NDFFL A L DF E+ARL IFE FCRIHQCI+I MLA +LNM+ EEAERWIV+LIR R
Sbjct 318 SNDFFLTAVLGDFRESARLLIFEMFCRIHQCITIEMLARRLNMSQEEAERWIVDLIRTYR 377
Query 121 LD-AKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEK 165
++ AKIDSKLG VVMG +VS ++QV+E TK L+ R+Q +A+ +EK
Sbjct 378 IEGAKIDSKLGQVVMGVKSVSIHEQVMENTKRLTLRAQQIALQLEK 423
> ath:AT3G57290 EIF3E; EIF3E (EUKARYOTIC TRANSLATION INITIATION
FACTOR 3E); translation initiation factor; K03250 translation
initiation factor 3 subunit E
Length=441
Score = 187 bits (474), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 94/170 (55%), Positives = 123/170 (72%), Gaps = 14/170 (8%)
Query 1 AVITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVL 60
A I NK +RR LK+ +KVIQQE Y+YKDPI EF+ C++VN+DFDGAQKK++ECE V+
Sbjct 261 AFIVNK---RRRPQLKEFIKVIQQEHYSYKDPIIEFLACVFVNYDFDGAQKKMKECEEVI 317
Query 61 VNDFFLVACLED-----------FIENARLFIFETFCRIHQCISINMLADKLNMTPEEAE 109
VND FL +ED F+ENARLF+FET+C+IHQ I + +LA+KLN+ EEAE
Sbjct 318 VNDPFLGKRVEDGNFSTVPLRDEFLENARLFVFETYCKIHQRIDMGVLAEKLNLNYEEAE 377
Query 110 RWIVNLIRNARLDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQML 159
RWIVNLIR ++LDAKIDS+ G V+M + ++Q+I TK LS R+ L
Sbjct 378 RWIVNLIRTSKLDAKIDSESGTVIMEPTQPNVHEQLINHTKGLSGRTYKL 427
> tgo:TGME49_073520 proteasome PCI domain-containing protein ;
K03250 translation initiation factor 3 subunit E
Length=601
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 76/168 (45%), Positives = 106/168 (63%), Gaps = 8/168 (4%)
Query 9 RKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKL----RECESVLVNDF 64
RKR+ + ++ I Q Y DP T +E L+++F+FD AQ + EC+S DF
Sbjct 362 RKRKDHFQAVITAIGQGKNRYSDPFTSLLEALFLDFNFDEAQHHIARIGEECDS----DF 417
Query 65 FLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAK 124
FL E ARL IFET+CRIH+CI+++M+A K+NM+PE AERWIVNLIR+ARL+A+
Sbjct 418 FLQPLKGAINEFARLLIFETYCRIHKCINVDMIAKKVNMSPEAAERWIVNLIRHARLEAR 477
Query 125 IDSKLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSR 172
IDS+ V M + YQQVI+KT++L RS ++ NI + R
Sbjct 478 IDSEKNRVQMSAAPPNLYQQVIDKTRNLCIRSNLILQNIARPAGFTGR 525
> cpv:cgd2_1500 possible translation initiation factor with possible
PINT domain ; K03250 translation initiation factor 3
subunit E
Length=540
Score = 138 bits (348), Expect = 9e-33, Method: Compositional matrix adjust.
Identities = 75/177 (42%), Positives = 108/177 (61%), Gaps = 3/177 (1%)
Query 8 VRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLV 67
+R+ + +L ++V +I YKD TE + L VNFDF+ +QK L SV NDFFL
Sbjct 350 MRRNKDILDNIVNIITLNRDKYKDLFTELIFNLLVNFDFEASQKILNSFSSVTENDFFLN 409
Query 68 ACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDS 127
+ ENARL IFE +CRIH+ I +N+++++L+MT EAERWIVNLIRN LD K+D
Sbjct 410 PLQKYIEENARLLIFEIYCRIHKSIDLNVISNELSMTSIEAERWIVNLIRNVHLDGKLDY 469
Query 128 KLGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLN---QNSRSEAPNWAAQ 181
+ V M + + +P Q V +KT+++ FRS +L N+ + NS+ NW Q
Sbjct 470 EHNRVEMIDRSNNPSQLVSDKTRNILFRSNLLIQNLSNNPSFNYNNSQKNKQNWRVQ 526
> tpv:TP02_0376 eukaryotic translation initiation factor 3 subunit
6; K03250 translation initiation factor 3 subunit E
Length=527
Score = 119 bits (299), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 61/157 (38%), Positives = 96/157 (61%), Gaps = 0/157 (0%)
Query 9 RKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVA 68
R R+ K + VI + Y D T + L+V+F+F+ AQK + E + D L
Sbjct 317 RNRKDHFKTISNVIANTKHKYNDTFTSLLGALFVDFNFEAAQKHIVEIKEACYVDVLLNP 376
Query 69 CLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSK 128
+ E++R IFET+CRIH+ I+++++A +NMT +AERWIVN+IR++ ++AKIDS+
Sbjct 377 LKDAIEESSRHIIFETYCRIHKSINLDIIAQNVNMTSLDAERWIVNIIRHSHVEAKIDSE 436
Query 129 LGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEK 165
V + + YQQVIEKT++L+ RS M+ N +
Sbjct 437 KNCVEISTVPPNLYQQVIEKTQNLTLRSNMILQNFSQ 473
> bbo:BBOV_III005760 17.m07513; eukaryotic translation initiation
factor 3, subunit 6; K03250 translation initiation factor
3 subunit E
Length=515
Score = 116 bits (291), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 66/176 (37%), Positives = 100/176 (56%), Gaps = 3/176 (1%)
Query 9 RKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVA 68
R R+ K + I Y D T V L+V+F+F+ AQK + + D FL
Sbjct 311 RNRKDHFKVISAAISNGKSKYSDTFTALVGALFVDFNFEDAQKHIARIQDACRLDVFLDP 370
Query 69 CLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSK 128
E++R IFET+CRIH+ I+++++A K+NM+ EAERWIV LIR++ L+AKIDS+
Sbjct 371 LKFQIEEHSRHIIFETYCRIHKSINLDLIAQKVNMSALEAERWIVGLIRHSHLEAKIDSE 430
Query 129 LGHVVMGNNAVSPYQQVIEKTKSLSFRSQMLAMNIEKKLNQNSRSEAPNWAAQDSG 184
V + + YQQVIEKT++L+ RS ++ N+ +N +S + D G
Sbjct 431 KNCVEISTVPPNLYQQVIEKTQNLTLRSNIILQNL---INADSSGPVYTRSQDDKG 483
> pfa:PFE1405c eukaryotic translation initiation factor 3, subunit
6, putative; K03250 translation initiation factor 3 subunit
E
Length=517
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 56/149 (37%), Positives = 90/149 (60%), Gaps = 0/149 (0%)
Query 29 YKDPITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENARLFIFETFCRI 88
Y D T + ++ ++DF+ AQ + ++ D FL E+ R IFET+CRI
Sbjct 334 YNDSFTSLLISIFTDYDFNMAQTCIATIGTLCERDVFLYKLKPYIEEHCRFIIFETYCRI 393
Query 89 HQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVMGNNAVSPYQQVIEK 148
H+ I+I+M++ K+N+T E+AE+WIVNLIRNA+LDAKI+S+ + + + YQQVI+K
Sbjct 394 HKSINIDMISSKVNLTREQAEKWIVNLIRNAKLDAKINSEKNCIEISTPPPNLYQQVIDK 453
Query 149 TKSLSFRSQMLAMNIEKKLNQNSRSEAPN 177
T++L RS + + + N + + N
Sbjct 454 TQNLIMRSNFIVQVLNRSSNDDQQQIQKN 482
> tpv:TP04_0395 26S proteasome regulatory subunit; K03037 26S
proteasome regulatory subunit N7
Length=393
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/130 (26%), Positives = 64/130 (49%), Gaps = 7/130 (5%)
Query 33 ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCI 92
I+++ C Y N+ K L + +++ D +L F+ ARL ++ F R ++ +
Sbjct 264 ISDYYNCNYKNY-----MKHLVDVSKLILKDRYLGRHCRYFVRQARLPAYKQFLRPYKSV 318
Query 93 SINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVV--MGNNAVSPYQQVIEKTK 150
++ +AD ++ E E +V+ I RLD KID G + +G+ + Y + I++
Sbjct 319 TLKNMADAFQVSTEFIEEELVSYISGMRLDCKIDKVNGIIENNVGDERNNNYVKTIKQGD 378
Query 151 SLSFRSQMLA 160
L R Q L+
Sbjct 379 LLINRIQKLS 388
> tpv:TP04_0396 hypothetical protein; K15030 translation initiation
factor 3 subunit M
Length=450
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 73 FIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHV 132
+E L T C+ I I M+ L + PEEAE+ IVN I ++A ID V
Sbjct 347 LVEKLTLLTISTMCQQQSEIPIEMIEKNLQLPPEEAEQMIVNAINKGVMEALIDQNSKKV 406
Query 133 VMGN 136
++ +
Sbjct 407 IINH 410
> bbo:BBOV_II001850 18.m06141; PCI domain containing 26S proteasome
non-ATPase regulatory subunit 6 protein; K03037 26S proteasome
regulatory subunit N7
Length=396
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 65/132 (49%), Gaps = 11/132 (8%)
Query 33 ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCI 92
+ +F C Y N+ F+ A+ +++ D ++ + + ARL + F R ++ +
Sbjct 267 LYDFYHCNYKNYMFNLAR-----TYDLILRDKYMSRHCKYILRQARLPAYRQFLRPYKSV 321
Query 93 SINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVMGNNAV----SPYQQVIEK 148
+I ++ + P+ E +V+ I RLD KID L + ++ NN + + Y +++++
Sbjct 322 TIENMSHAFQLPPDFIEDELVSYISGMRLDCKID--LVNGIIENNIMDERNTNYIEIVKE 379
Query 149 TKSLSFRSQMLA 160
L R Q L+
Sbjct 380 GDMLLNRIQKLS 391
> ath:AT2G26990 FUS12; FUS12 (FUSCA 12); K12176 COP9 signalosome
complex subunit 2
Length=439
Score = 36.6 bits (83), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 18/86 (20%), Positives = 44/86 (51%), Gaps = 0/86 (0%)
Query 53 LRECESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWI 112
L+ +++D F+ +ED ++ R + + + I I ++ +LN+ + +
Sbjct 326 LKSNRRTIMDDPFIRNYMEDLLKKVRTQVLLKLIKPYTKIGIPFISKELNVPETDVTELL 385
Query 113 VNLIRNARLDAKIDSKLGHVVMGNNA 138
V+LI ++R+D ID +++ G++
Sbjct 386 VSLILDSRIDGHIDEMNRYLLRGDSG 411
> dre:723998 sb:cb336; zgc:136894
Length=497
Score = 34.3 bits (77), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Query 57 ESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPE 106
ES L CL DF + L TFCR QCIS + + +L + PE
Sbjct 2 ESRLSKQIQCSVCLGDFTDPVSLLCDHTFCR--QCISNHSQSSRLRLCPE 49
> xla:432342 cops2, csn2; COP9 constitutive photomorphogenic homolog
subunit 2; K12176 COP9 signalosome complex subunit 2
Length=443
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 52/108 (48%), Gaps = 6/108 (5%)
Query 24 QESYTYK-DP----ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENAR 78
QE+ YK DP +T V Y N D +K L+ S +++D F+ +E+ + N R
Sbjct 297 QEAKPYKNDPEILAMTNLVSA-YQNNDITEFEKILKTNHSNIMDDPFIREHIEELLRNIR 355
Query 79 LFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKID 126
+ + + I I ++ +LN+ + E +V I + ++ +ID
Sbjct 356 TQVLIKLIKPYTRIHIPFISKELNIDVADVESLLVQCILDNTINGRID 403
> dre:415145 cops2, wu:fc05f05, wu:fc06g10, wu:fc60f04, wu:fl94f12,
zgc:86624; COP9 constitutive photomorphogenic homolog
subunit 2 (Arabidopsis); K12176 COP9 signalosome complex subunit
2
Length=443
Score = 33.9 bits (76), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 52/108 (48%), Gaps = 6/108 (5%)
Query 24 QESYTYK-DP----ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENAR 78
QE+ YK DP +T V Y N D +K L+ S +++D F+ +E+ + N R
Sbjct 297 QEAKPYKNDPEILAMTNLVSA-YQNNDITEFEKILKTNHSNIMDDPFIREHIEELLRNIR 355
Query 79 LFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKID 126
+ + + I I ++ +LN+ + E +V I + ++ +ID
Sbjct 356 TQVLIKLIKPYTRIHIPFISKELNIDVADVESLLVQCILDNTINGRID 403
> bbo:BBOV_III007800 17.m07684; heat shock protein 70 precursor;
K09490 heat shock 70kDa protein 5
Length=652
Score = 33.1 bits (74), Expect = 0.50, Method: Composition-based stats.
Identities = 28/92 (30%), Positives = 43/92 (46%), Gaps = 10/92 (10%)
Query 93 SINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVMGNNAVSPYQQVIEKTKSL 152
+I + DK ++PEE ER +IR+A +A+ D ++ V A+ Y ++KT
Sbjct 525 NIVITPDKGRLSPEEIER----MIRDAEANAEKDKEVFEKVQSKQALEVYLDSMKKTVQ- 579
Query 153 SFRSQMLAMNIE---KKLNQNSRSEAPNWAAQ 181
LA IE K + + SEA W Q
Sbjct 580 --DKDKLAHKIEDDDKDIIMKAISEAETWMMQ 609
> mmu:12848 Cops2, AI315723, C85265, Csn2, Sgn2, Trip15, alien-like;
COP9 (constitutive photomorphogenic) homolog, subunit
2 (Arabidopsis thaliana); K12176 COP9 signalosome complex subunit
2
Length=443
Score = 33.1 bits (74), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 6/108 (5%)
Query 24 QESYTYK-DP----ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENAR 78
QE+ YK DP +T V Y N D +K L+ S +++D F+ +E+ + N R
Sbjct 297 QEAKPYKNDPEILAMTNLVSA-YQNNDITEFEKILKTNHSNIMDDPFIREHIEELLRNIR 355
Query 79 LFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKID 126
+ + + I I ++ +LN+ + E +V I + + +ID
Sbjct 356 TQVLIKLIKPYTRIHIPFISKELNIDVADVESLLVQCILDNTIHGRID 403
> hsa:9318 COPS2, ALIEN, CSN2, SGN2, TRIP15; COP9 constitutive
photomorphogenic homolog subunit 2 (Arabidopsis); K12176 COP9
signalosome complex subunit 2
Length=450
Score = 33.1 bits (74), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 6/108 (5%)
Query 24 QESYTYK-DP----ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENAR 78
QE+ YK DP +T V Y N D +K L+ S +++D F+ +E+ + N R
Sbjct 304 QEAKPYKNDPEILAMTNLVSA-YQNNDITEFEKILKTNHSNIMDDPFIREHIEELLRNIR 362
Query 79 LFIFETFCRIHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKID 126
+ + + I I ++ +LN+ + E +V I + + +ID
Sbjct 363 TQVLIKLIKPYTRIHIPFISKELNIDVADVESLLVQCILDNTIHGRID 410
> tgo:TGME49_061210 26S proteasome non-ATPase regulatory subunit,
putative ; K03035 26S proteasome regulatory subunit N5
Length=485
Score = 32.7 bits (73), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 89 HQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVMGN 136
+ CI ++ +A L++T +EAE I L+ + ++AKID G V G
Sbjct 387 YSCIEMDRVASLLDITKDEAESEISELVCSNFIEAKIDRPAGTVEFGK 434
> cpv:cgd2_2210 proteasome regulatory subunit Rpn7/26S proteasome
subunit 6, PINT domain containing protein ; K03037 26S proteasome
regulatory subunit N7
Length=403
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 29/135 (21%), Positives = 63/135 (46%), Gaps = 5/135 (3%)
Query 33 ITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCI 92
+ EF+E Y + ++L +L D++ + ++ R + + ++ +
Sbjct 270 LLEFIESFYYG-RYSQFFERLVNITYILQKDYYFNRHHKYYLRIVRSKAYIQYLEPYESV 328
Query 93 SINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVM---GNNAVSPYQQVIEKT 149
SI +A+ +T E E+ IV I +++L ID K+ V++ + ++ Y ++++K
Sbjct 329 SILSMAESFGITQEFLEKDIVTFISSSKLSCTID-KVKQVIICSRNDKKINQYNELVQKG 387
Query 150 KSLSFRSQMLAMNIE 164
L R Q L I+
Sbjct 388 DLLLNRLQRLTRIIQ 402
> dre:335720 cops3, wu:fc32a02, wu:fk59f01, zgc:111903, zgc:55643,
zgc:77615; COP9 constitutive photomorphogenic homolog subunit
3; K12177 COP9 signalosome complex subunit 3
Length=423
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 92 ISINMLADKLNMT-PEEAERWIVNLIRNARLDAKIDSKLGHVVMGNN 137
+S+ +A ++ ++ P+EAE++++++I + + A I+ K G V +N
Sbjct 317 LSLQDMASRVQLSGPQEAEKYVLHMIEDGEIYASINQKDGMVCFHDN 363
> dre:100334543 COP9 signalosome complex subunit 3-like
Length=418
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 92 ISINMLADKLNMT-PEEAERWIVNLIRNARLDAKIDSKLGHVVMGNN 137
+S+ +A ++ ++ P+EAE++++++I + + A I+ K G V +N
Sbjct 339 LSLQDMASRVQLSGPQEAEKYVLHMIEDGEIYASINQKDGMVCFHDN 385
> mmu:26572 Cops3, Csn3, Sgn3; COP9 (constitutive photomorphogenic)
homolog, subunit 3 (Arabidopsis thaliana); K12177 COP9
signalosome complex subunit 3
Length=423
Score = 30.4 bits (67), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 92 ISINMLADKLNMT-PEEAERWIVNLIRNARLDAKIDSKLGHVVMGNN 137
+S+ +A ++ ++ P+EAE++++++I + + A I+ K G V +N
Sbjct 317 LSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDN 363
> hsa:8533 COPS3, CSN3, SGN3; COP9 constitutive photomorphogenic
homolog subunit 3 (Arabidopsis); K12177 COP9 signalosome
complex subunit 3
Length=403
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 92 ISINMLADKLNMT-PEEAERWIVNLIRNARLDAKIDSKLGHVVMGNN 137
+S+ +A ++ ++ P+EAE++++++I + + A I+ K G V +N
Sbjct 297 LSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDN 343
> cel:W02D3.8 smg-5; Suppressor with Morphological effect on Genitalia
family member (smg-5)
Length=549
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 14/67 (20%), Positives = 35/67 (52%), Gaps = 2/67 (2%)
Query 88 IHQCISINMLADKLNMTPEEAERWIVNLIRNARLDAKIDSKLGHVVMGNNAVSPYQQVIE 147
I +C+++ + +++ P + + +++ R D + KL +++G A +PY++ E
Sbjct 124 IQKCLAVGYYSRAIDLDPNQGRAF--HVLAGLRADLNVAQKLRLMILGQLADAPYKKGTE 181
Query 148 KTKSLSF 154
+ L F
Sbjct 182 LLEYLKF 188
> cel:Y48G10A.4 hypothetical protein; K13145 integrator complex
subunit 8
Length=965
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 2/53 (3%)
Query 3 ITNKDVRKRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRE 55
I+N D RQ+L+ +V+ S T D I + + LYV DF GA +K E
Sbjct 769 ISNPD--DVRQLLQLMVEKAYSLSPTDPDTIRTYADFLYVERDFQGAAQKYME 819
> pfa:PFF1440w SET domain protein, putative
Length=6753
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 47/89 (52%), Gaps = 6/89 (6%)
Query 10 KRRQVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVAC 69
K+R+ +++++ V+ +SY + +E++ LY + ++D A+KK + +L ND C
Sbjct 2456 KKRKSVEEIMSVVDNKSYYGFNKCSEYI--LYSSNEYDRAKKKENKRIKLLKNDILKECC 2513
Query 70 -LEDFIENARLFIFETFCRI---HQCISI 94
+ IE FI+ C I + C +I
Sbjct 2514 YICGSIEYKNNFIYCCICGISVHYSCANI 2542
> tpv:TP03_0646 hypothetical protein
Length=467
Score = 29.6 bits (65), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 61 VNDFFLVACLEDFIENA-RLFIFETFCRIHQCISINMLADKLNMTPEE 107
+ F V L EN R+F T CR +SI +L D LN + E+
Sbjct 381 TDSMFYVKLLFKMFENKLRMFSLYTLCRSSLTLSIELLTDILNFSSED 428
> sce:YJR049C UTR1; ATP-NADH kinase; phosphorylates both NAD and
NADH; active as a hexamer; enhances the activity of ferric
reductase (Fre1p) (EC:2.7.1.23)
Length=530
Score = 29.6 bits (65), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 26/53 (49%), Gaps = 4/53 (7%)
Query 135 GNNAVSPYQQVIEKTK----SLSFRSQMLAMNIEKKLNQNSRSEAPNWAAQDS 183
G + V P I T +LSFR +L +I K+ + +S AP WAA D
Sbjct 350 GGSLVCPTVNAIALTPICPHALSFRPIILPESINLKVKVSMKSRAPAWAAFDG 402
> mmu:66763 4933425L06Rik, MGC141320; RIKEN cDNA 4933425L06 gene
Length=585
Score = 29.3 bits (64), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 32/69 (46%), Gaps = 6/69 (8%)
Query 55 ECESVLVNDFFLVACLEDFIENARLFIFETFCRIHQCISINMLADKLNMTPEEAERWIVN 114
E S L D ++ A + DF +N + + E C I++ D +T E + N
Sbjct 285 EILSYLEEDSYIKAIVRDFTQNIQYMLEEVLC------PIDVALDGREITVRRCESNLGN 338
Query 115 LIRNARLDA 123
L+ NA L+A
Sbjct 339 LVTNAMLEA 347
> ath:AT1G64990 GTG1; GTG1 (GPCR-TYPE G PROTEIN 1)
Length=468
Score = 28.9 bits (63), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 1/70 (1%)
Query 13 QVLKDLVKVIQQESYTYKDPITEFVECLYVNFDFDGAQKKLRECESVLVNDFFLVACLED 72
++LK L V+ +E+ T KDP+T + FD L + S+L +V +
Sbjct 309 KMLKSLQSVVFKEAGT-KDPVTTMISIFLRLFDIGVDAALLSQYISLLFIGMLIVISVRG 367
Query 73 FIENARLFIF 82
F+ N F F
Sbjct 368 FLTNLMKFFF 377
Lambda K H
0.322 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40