bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3757_orf1
Length=103
Score E
Sequences producing significant alignments: (Bits) Value
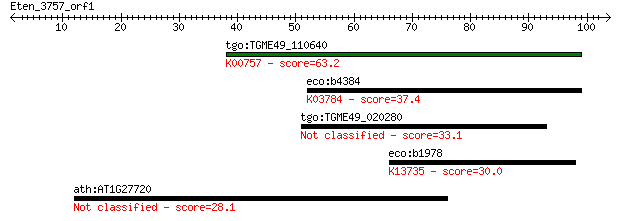
tgo:TGME49_110640 uridine phosphorylase, putative (EC:2.4.2.1)... 63.2 2e-10
eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosph... 37.4 0.011
tgo:TGME49_020280 SCP-like domain-containing protein 33.1 0.24
eco:b1978 yeeJ, ECK1974, JW5833; probable adhesin; K13735 adhe... 30.0 1.9
ath:AT1G27720 TAF4B; TAF4B (TBP-ASSOCIATED FACTOR 4B); transcr... 28.1 8.0
> tgo:TGME49_110640 uridine phosphorylase, putative (EC:2.4.2.1);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=303
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 38/61 (62%), Gaps = 0/61 (0%)
Query 38 KEAEIFLNEDGSLYHLGVKSGELHPRILTVGDRERAQMIAEAFLEQVREFEKSRNFRTFS 97
K IFL DG YHLGVK G+L I+TVG +RA+++A FL V E SR F TF+
Sbjct 5 KGKNIFLTPDGRTYHLGVKKGDLASLIVTVGCEQRARVLAAKFLTDVTEVSSSRQFYTFT 64
Query 98 G 98
G
Sbjct 65 G 65
> eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosphorylase
(EC:2.4.2.1); K03784 purine-nucleoside phosphorylase
[EC:2.4.2.1]
Length=239
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 52 HLGVKSGELHPRILTVGDRERAQMIAEAFLEQVREFEKSRNFRTFSG 98
H+ + G+ +L GD RA+ IAE FLE RE R F+G
Sbjct 5 HINAEMGDFADVVLMPGDPLRAKYIAETFLEDAREVNNVRGMLGFTG 51
> tgo:TGME49_020280 SCP-like domain-containing protein
Length=434
Score = 33.1 bits (74), Expect = 0.24, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 27/48 (56%), Gaps = 6/48 (12%)
Query 51 YHLGVKSGELHPRI--LTVGDRERAQMIAEAFL----EQVREFEKSRN 92
YH + +L PR+ LT GD+ERA +AF+ E V E ++ RN
Sbjct 62 YHDAKRGRQLSPRLSELTTGDKERASQRLQAFIDNGCEDVVEVDECRN 109
> eco:b1978 yeeJ, ECK1974, JW5833; probable adhesin; K13735 adhesin/invasin
Length=2358
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 66 TVGDRERAQMIAEAFLEQVREFEKSRNFRTFS 97
T+G E AQ +AE F V E K FRTF+
Sbjct 72 TLGALESAQSVAERFGISVAELRKLNQFRTFA 103
> ath:AT1G27720 TAF4B; TAF4B (TBP-ASSOCIATED FACTOR 4B); transcription
initiation factor
Length=720
Score = 28.1 bits (61), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 32/68 (47%), Gaps = 4/68 (5%)
Query 12 RGVHTSQHCAGFAKTPTSSFSQKMAEKEAEIFLNEDGSLYHLGVKSGELHP----RILTV 67
R VH +Q + + T SS + + K E + S +H+ KSG L+P + +
Sbjct 178 REVHVNQLSSTTSGTLNSSTTVQGLNKHPEQHMQLPSSSFHMDTKSGSLNPYPGTNVTSP 237
Query 68 GDRERAQM 75
G RA++
Sbjct 238 GSSSRAKL 245
Lambda K H
0.321 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027061836
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40