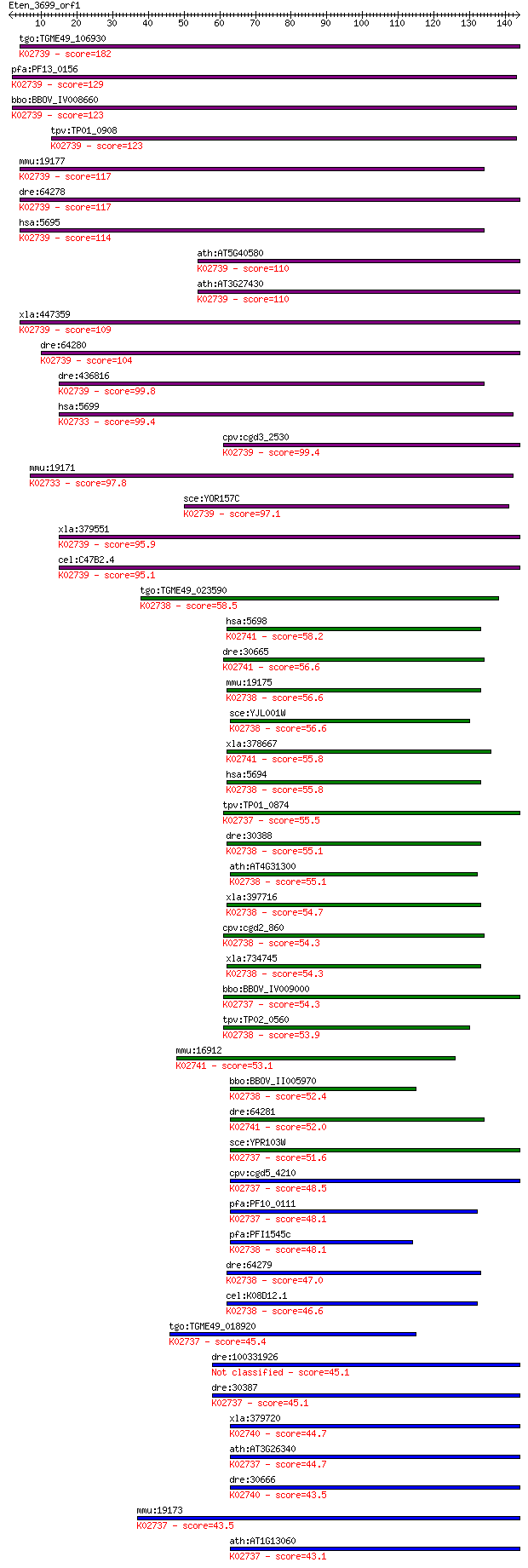
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3699_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_106930 proteasome subunit beta type 7, putative (EC... 182 2e-46
pfa:PF13_0156 proteasome subunit beta type 7 precursor, putati... 129 4e-30
bbo:BBOV_IV008660 23.m05823; proteasome subunit beta 7 (EC:3.4... 123 2e-28
tpv:TP01_0908 proteasome subunit beta type 7 precursor; K02739... 123 2e-28
mmu:19177 Psmb7, AU020723, MC14; proteasome (prosome, macropai... 117 8e-27
dre:64278 psmb7, C167P9.3, MGC110090, unp311, zgc:110090; prot... 117 1e-26
hsa:5695 PSMB7, Z; proteasome (prosome, macropain) subunit, be... 114 1e-25
ath:AT5G40580 PBB2; PBB2; endopeptidase/ peptidase/ threonine-... 110 1e-24
ath:AT3G27430 PBB1; PBB1; endopeptidase/ peptidase/ threonine-... 110 1e-24
xla:447359 psmb7, MGC84123; proteasome (prosome, macropain) su... 109 3e-24
dre:64280 psmb10, psmb12, zgc:110363; proteasome (prosome, mac... 104 9e-23
dre:436816 zgc:92791 (EC:3.4.25.1); K02739 20S proteasome subu... 99.8 2e-21
hsa:5699 PSMB10, FLJ00366, LMP10, MECL1, MGC1665, beta2i; prot... 99.4 3e-21
cpv:cgd3_2530 PUP1/proteasome subunit beta type 7, NTN hydrola... 99.4 3e-21
mmu:19171 Psmb10, Mecl1; proteasome (prosome, macropain) subun... 97.8 9e-21
sce:YOR157C PUP1; Beta 2 subunit of the 20S proteasome; endope... 97.1 2e-20
xla:379551 psmb7.2, MGC68991; proteasome (prosome, macropain) ... 95.9 4e-20
cel:C47B2.4 pbs-2; Proteasome Beta Subunit family member (pbs-... 95.1 7e-20
tgo:TGME49_023590 proteasome component PRE3 precursor, putativ... 58.5 6e-09
hsa:5698 PSMB9, LMP2, MGC70470, PSMB6i, RING12, beta1i; protea... 58.2 9e-09
dre:30665 psmb9a, lmp2, macropain, prosome, psmb9, wu:fa66f04,... 56.6 2e-08
mmu:19175 Psmb6, Lmp19, Mpnd; proteasome (prosome, macropain) ... 56.6 3e-08
sce:YJL001W PRE3, CRL21; Beta 1 subunit of the 20S proteasome,... 56.6 3e-08
xla:378667 psmb9, LMP2, psmb9-A; proteasome (prosome, macropai... 55.8 4e-08
hsa:5694 PSMB6, DELTA, LMPY, MGC5169, Y; proteasome (prosome, ... 55.8 5e-08
tpv:TP01_0874 proteasome subunit beta type 5; K02737 20S prote... 55.5 5e-08
dre:30388 psmb6, y, zgc:109823; proteasome (prosome, macropain... 55.1 7e-08
ath:AT4G31300 PBA1; PBA1; endopeptidase/ peptidase/ threonine-... 55.1 8e-08
xla:397716 proteasome subunit Y; K02738 20S proteasome subunit... 54.7 1e-07
cpv:cgd2_860 Pre3p/proteasome regulatory subunit beta type 6, ... 54.3 1e-07
xla:734745 hypothetical protein MGC130855; K02738 20S proteaso... 54.3 1e-07
bbo:BBOV_IV009000 23.m05743; proteasome epsilon subunit; K0273... 54.3 1e-07
tpv:TP02_0560 proteasome precursor; K02738 20S proteasome subu... 53.9 2e-07
mmu:16912 Psmb9, Lmp-2, Lmp2; proteasome (prosome, macropain) ... 53.1 2e-07
bbo:BBOV_II005970 18.m06495; proteosome A; K02738 20S proteaso... 52.4 5e-07
dre:64281 psmb9b; proteasome (prosome, macropain) subunit, bet... 52.0 6e-07
sce:YPR103W PRE2, DOA3, PRG1, SRR2; Beta 5 subunit of the 20S ... 51.6 8e-07
cpv:cgd5_4210 Pre2p/proteasome subunit beta type 5; NTN hydrol... 48.5 7e-06
pfa:PF10_0111 20S proteasome beta subunit, putative; K02737 20... 48.1 8e-06
pfa:PFI1545c proteasome precursor, putative (EC:3.4.25.1); K02... 48.1 9e-06
dre:64279 psmb11, MGC92422, macropain, prosome, zgc:92422; pro... 47.0 2e-05
cel:K08D12.1 pbs-1; Proteasome Beta Subunit family member (pbs... 46.6 3e-05
tgo:TGME49_018920 proteasome subunit beta type 5, putative (EC... 45.4 5e-05
dre:100331926 proteasome (prosome, macropain) subunit, beta ty... 45.1 8e-05
dre:30387 psmb5, etID309919.2, fb59c07, si:zc14a17.13, wu:fb59... 45.1 8e-05
xla:379720 MGC69086; low molecular mass protein-7 (LMP-7) homo... 44.7 9e-05
ath:AT3G26340 20S proteasome beta subunit E, putative; K02737 ... 44.7 1e-04
dre:30666 psmb8, lmp7, macropain, prosome; proteasome (prosome... 43.5 2e-04
mmu:19173 Psmb5, MGC118075; proteasome (prosome, macropain) su... 43.5 2e-04
ath:AT1G13060 PBE1; PBE1; endopeptidase/ peptidase/ threonine-... 43.1 3e-04
> tgo:TGME49_106930 proteasome subunit beta type 7, putative (EC:3.4.25.1);
K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=368
Score = 182 bits (463), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 91/140 (65%), Positives = 106/140 (75%), Gaps = 9/140 (6%)
Query 4 MGSVEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTG 63
M S+ QL Q GG+DFSNH RNV LL+ +A + + S P +PP RKTG
Sbjct 83 MDSISQLALQRGGYDFSNHQRNVRLLMEAAKRCPAVAGSPTPPG--------LPPARKTG 134
Query 64 TTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLA 123
TTICGV+ +DGVVLGADTRATEG +VADKNC+KLH+I+ N+YAAGAGT+ADLDHMCDWLA
Sbjct 135 TTICGVVCKDGVVLGADTRATEGTIVADKNCSKLHRIADNMYAAGAGTSADLDHMCDWLA 194
Query 124 TQVELHRLNTYAKQARVSMA 143
QVELHRLNT AK RVSMA
Sbjct 195 VQVELHRLNTNAK-PRVSMA 213
> pfa:PF13_0156 proteasome subunit beta type 7 precursor, putative
(EC:3.4.25.1); K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=270
Score = 129 bits (323), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 70/141 (49%), Positives = 85/141 (60%), Gaps = 22/141 (15%)
Query 2 FKMGSVEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRK 61
K+ + L+ + GG++F N RN L GV + P RK
Sbjct 1 MKLEYINILKEENGGYNFDNLKRNEIL------------KEKGV---------KFPQFRK 39
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTICG++ Q+ V+LGADTRATEGP+VADKNC+KLH IS NI+ AGAG A DL+H W
Sbjct 40 TGTTICGLVCQNAVILGADTRATEGPIVADKNCSKLHYISKNIWCAGAGVAGDLEHTTLW 99
Query 122 LATQVELHRLNTYAKQARVSM 142
L VELHRLNT Q RVSM
Sbjct 100 LQHNVELHRLNT-NTQPRVSM 119
> bbo:BBOV_IV008660 23.m05823; proteasome subunit beta 7 (EC:3.4.25.1);
K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=271
Score = 123 bits (309), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 69/144 (47%), Positives = 82/144 (56%), Gaps = 25/144 (17%)
Query 2 FKMGSVEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRK 61
+ G V + GG+DFSNH RN A + + + K
Sbjct 1 MEFGYVREYMSHQGGWDFSNHSRN----------------------ARIKDNIKGFKVLK 38
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTICGV+ +DGVVL ADTRATEGP+VADKNC+KLH+IS IY AGAG AADL+H W
Sbjct 39 TGTTICGVLVKDGVVLAADTRATEGPIVADKNCSKLHRISDFIYCAGAGVAADLEHTTLW 98
Query 122 LATQVELHRLNTYAK---QARVSM 142
L +EL RLN K Q VSM
Sbjct 99 LENNIELLRLNLKQKPKVQMCVSM 122
> tpv:TP01_0908 proteasome subunit beta type 7 precursor; K02739
20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=262
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 68/130 (52%), Positives = 78/130 (60%), Gaps = 23/130 (17%)
Query 13 QLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTGTTICGVIFQ 72
Q GG+DFSN+ RN + S KTGTTICGV+ +
Sbjct 3 QKGGWDFSNYSRNSRI----------------------QNSINGHNYLKTGTTICGVMGK 40
Query 73 DGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLATQVELHRLN 132
D VVL ADTRAT+G +VADKNC+KLHKIS NIY AGAG AADL+H WLA +ELHRLN
Sbjct 41 DSVVLAADTRATQGIIVADKNCSKLHKISDNIYCAGAGVAADLEHTTLWLANNIELHRLN 100
Query 133 TYAKQARVSM 142
T KQ R M
Sbjct 101 T-KKQPRAQM 109
> mmu:19177 Psmb7, AU020723, MC14; proteasome (prosome, macropain)
subunit, beta type 7 (EC:3.4.25.1); K02739 20S proteasome
subunit beta 2 [EC:3.4.25.1]
Length=277
Score = 117 bits (294), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 62/130 (47%), Positives = 78/130 (60%), Gaps = 17/130 (13%)
Query 4 MGSVEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTG 63
M +V Q +GGF F N RN L + A F++P RKTG
Sbjct 1 MAAVSVFQPPVGGFSFDNCRRNAVL-----------------EADFAKKGFKLPKARKTG 43
Query 64 TTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLA 123
TTI GV+++DG+VLGADTRATEG +VADKNC+K+H IS NIY GAGTAAD D ++
Sbjct 44 TTIAGVVYKDGIVLGADTRATEGMVVADKNCSKIHFISPNIYCCGAGTAADTDMTTQLIS 103
Query 124 TQVELHRLNT 133
+ +ELH L T
Sbjct 104 SNLELHSLTT 113
> dre:64278 psmb7, C167P9.3, MGC110090, unp311, zgc:110090; proteasome
(prosome, macropain) subunit, beta type, 7 (EC:3.4.25.1);
K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=277
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 66/140 (47%), Positives = 84/140 (60%), Gaps = 18/140 (12%)
Query 4 MGSVEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTG 63
M +V Q Q GGF F N CR +LL + F P RKTG
Sbjct 1 MATVSVCQYQPGGFSFEN-CRRNALL----------------EADITKLGFSSPAARKTG 43
Query 64 TTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLA 123
TTICG++++DGVVLGADTRATEG +VADKNC+K+H IS NIY GAGTAAD + ++
Sbjct 44 TTICGIVYKDGVVLGADTRATEGMIVADKNCSKIHYISPNIYCCGAGTAADTEMTTQIIS 103
Query 124 TQVELHRLNTYAKQARVSMA 143
+ +ELH L+T + RV+ A
Sbjct 104 SNLELHSLST-GRLPRVATA 122
> hsa:5695 PSMB7, Z; proteasome (prosome, macropain) subunit,
beta type, 7 (EC:3.4.25.1); K02739 20S proteasome subunit beta
2 [EC:3.4.25.1]
Length=277
Score = 114 bits (285), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 60/130 (46%), Positives = 79/130 (60%), Gaps = 17/130 (13%)
Query 4 MGSVEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTG 63
M +V +GGF F N RN L + A +++P +RKTG
Sbjct 1 MAAVSVYAPPVGGFSFDNCRRNAVL-----------------EADFAKRGYKLPKVRKTG 43
Query 64 TTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLA 123
TTI GV+++DG+VLGADTRATEG +VADKNC+K+H IS NIY GAGTAAD D ++
Sbjct 44 TTIAGVVYKDGIVLGADTRATEGMVVADKNCSKIHFISPNIYCCGAGTAADTDMTTQLIS 103
Query 124 TQVELHRLNT 133
+ +ELH L+T
Sbjct 104 SNLELHSLST 113
> ath:AT5G40580 PBB2; PBB2; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=274
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 51/90 (56%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 54 FRMPPLRKTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAA 113
+ P KTGTTI G+IF+DGV+LGADTRATEGP+VADKNC K+H ++ NIY GAGTAA
Sbjct 30 LKAPSFLKTGTTIVGLIFKDGVILGADTRATEGPIVADKNCEKIHYMAPNIYCCGAGTAA 89
Query 114 DLDHMCDWLATQVELHRLNTYAKQARVSMA 143
D + + D +++Q+ LHR T + +RV A
Sbjct 90 DTEAVTDMVSSQLRLHRYQT-GRDSRVVTA 118
> ath:AT3G27430 PBB1; PBB1; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=273
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 51/90 (56%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 54 FRMPPLRKTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAA 113
+ P KTGTTI G+IF+DGV+LGADTRATEGP+VADKNC K+H ++ NIY GAGTAA
Sbjct 30 LKAPSFLKTGTTIVGLIFKDGVILGADTRATEGPIVADKNCEKIHYMAPNIYCCGAGTAA 89
Query 114 DLDHMCDWLATQVELHRLNTYAKQARVSMA 143
D + + D +++Q+ LHR T + +RV A
Sbjct 90 DTEAVTDMVSSQLRLHRYQT-GRDSRVITA 118
> xla:447359 psmb7, MGC84123; proteasome (prosome, macropain)
subunit, beta type, 7 (EC:3.4.25.1); K02739 20S proteasome subunit
beta 2 [EC:3.4.25.1]
Length=277
Score = 109 bits (273), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 60/140 (42%), Positives = 82/140 (58%), Gaps = 18/140 (12%)
Query 4 MGSVEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTG 63
M +V + GFDF N CR +LL ++P RKTG
Sbjct 1 MATVTVRSAPVAGFDFGN-CRRNTLLTNEFTQKGH----------------KLPSSRKTG 43
Query 64 TTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLA 123
TTI G++F+DGV+LGADTRATEG +VADKNC+K+H I+ N+Y GAGTAAD + ++
Sbjct 44 TTIAGIVFKDGVILGADTRATEGMIVADKNCSKIHYIAPNVYCCGAGTAADTEMTTQMIS 103
Query 124 TQVELHRLNTYAKQARVSMA 143
+ +ELH L+T + RV+ A
Sbjct 104 SNMELHSLST-GRLPRVATA 122
> dre:64280 psmb10, psmb12, zgc:110363; proteasome (prosome, macropain)
subunit, beta type, 10 (EC:3.4.25.1); K02739 20S proteasome
subunit beta 2 [EC:3.4.25.1]
Length=281
Score = 104 bits (260), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 76/134 (56%), Gaps = 18/134 (13%)
Query 10 LQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTGTTICGV 69
L+ L GF+F N RN+ L + A G + P KTGTTI GV
Sbjct 8 LEPSLCGFNFENATRNIVL-----------------ENGAEEGKIKPPKALKTGTTIAGV 50
Query 70 IFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLATQVELH 129
+F+DGVVLGADTRAT +VADK C K+H I+ NIY GAGTAAD + D L++ + +
Sbjct 51 VFKDGVVLGADTRATSDEVVADKMCAKIHYIAPNIYCCGAGTAADTEKTTDMLSSNLTIF 110
Query 130 RLNTYAKQARVSMA 143
+N+ + RV MA
Sbjct 111 SMNS-GRNPRVVMA 123
> dre:436816 zgc:92791 (EC:3.4.25.1); K02739 20S proteasome subunit
beta 2 [EC:3.4.25.1]
Length=276
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 55/119 (46%), Positives = 69/119 (57%), Gaps = 17/119 (14%)
Query 15 GGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTGTTICGVIFQDG 74
GGF F N RN L + SA P RKTGTTI G++F+DG
Sbjct 11 GGFPFENTRRNAVLEANLSEKGYSA-----------------PNARKTGTTIAGLVFKDG 53
Query 75 VVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLATQVELHRLNT 133
V+LGADTRAT+ +VADKNC K+H I+ NIY GAG AAD + +++ VELH L+T
Sbjct 54 VILGADTRATDDMVVADKNCMKIHYIAPNIYCCGAGVAADAEVTTQMMSSSVELHSLST 112
> hsa:5699 PSMB10, FLJ00366, LMP10, MECL1, MGC1665, beta2i; proteasome
(prosome, macropain) subunit, beta type, 10 (EC:3.4.25.1);
K02733 20S proteasome subunit beta 10 [EC:3.4.25.1]
Length=273
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 55/127 (43%), Positives = 73/127 (57%), Gaps = 20/127 (15%)
Query 15 GGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTGTTICGVIFQDG 74
GGF F N RN SL ++P RKTGTTI G++FQDG
Sbjct 10 GGFSFENCQRNASL-------------------ERVLPGLKVPHARKTGTTIAGLVFQDG 50
Query 75 VVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLATQVELHRLNTY 134
V+LGADTRAT +VADK+C K+H I+ IY GAG AAD + +A+++ELH L+T
Sbjct 51 VILGADTRATNDSVVADKSCEKIHFIAPKIYCCGAGVAADAEMTTRMVASKMELHALST- 109
Query 135 AKQARVS 141
++ RV+
Sbjct 110 GREPRVA 116
> cpv:cgd3_2530 PUP1/proteasome subunit beta type 7, NTN hydrolase
fold ; K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=279
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 49/83 (59%), Positives = 62/83 (74%), Gaps = 1/83 (1%)
Query 61 KTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCD 120
KTGTTI GV D VVLGADTRAT GP+VADK+C K+H++S NI+AAGAGTAADLDH+
Sbjct 39 KTGTTIVGVACNDCVVLGADTRATNGPIVADKDCEKIHRLSDNIFAAGAGTAADLDHVTS 98
Query 121 WLATQVELHRLNTYAKQARVSMA 143
+ +EL +L ++ RV+ A
Sbjct 99 LIEGNLELQKLQM-NRKPRVAHA 120
> mmu:19171 Psmb10, Mecl1; proteasome (prosome, macropain) subunit,
beta type 10 (EC:3.4.25.1); K02733 20S proteasome subunit
beta 10 [EC:3.4.25.1]
Length=273
Score = 97.8 bits (242), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 56/135 (41%), Positives = 75/135 (55%), Gaps = 20/135 (14%)
Query 7 VEQLQRQLGGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTGTTI 66
++Q GGF F N RN SL R+P RKTGTTI
Sbjct 2 LKQAVEPTGGFSFENCQRNASL-------------------EHVLPGLRVPHARKTGTTI 42
Query 67 CGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLATQV 126
G++F+DGV+LGADTRAT +VADK+C K+H I+ IY GAG AAD + A+++
Sbjct 43 AGLVFRDGVILGADTRATNDSVVADKSCEKIHFIAPKIYCCGAGVAADTEMTTRMAASKM 102
Query 127 ELHRLNTYAKQARVS 141
ELH L+T ++ RV+
Sbjct 103 ELHALST-GREPRVA 116
> sce:YOR157C PUP1; Beta 2 subunit of the 20S proteasome; endopeptidase
with trypsin-like activity that cleaves after basic
residues; synthesized as a proprotein before being proteolytically
processed for assembly into 20S particle; human homolog
is subunit Z (EC:3.4.25.1); K02739 20S proteasome subunit
beta 2 [EC:3.4.25.1]
Length=261
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 48/91 (52%), Positives = 63/91 (69%), Gaps = 1/91 (1%)
Query 50 AAGSFRMPPLRKTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGA 109
A S P TGTTI GV F +GVV+ ADTR+T+GP+VADKNC KLH+IS I+ AGA
Sbjct 16 AENSHTQPKATSTGTTIVGVKFNNGVVIAADTRSTQGPIVADKNCAKLHRISPKIWCAGA 75
Query 110 GTAADLDHMCDWLATQVELHRLNTYAKQARV 140
GTAAD + + + + +ELH L T +++ RV
Sbjct 76 GTAADTEAVTQLIGSNIELHSLYT-SREPRV 105
> xla:379551 psmb7.2, MGC68991; proteasome (prosome, macropain)
subunit, beta type, 7, gene 2; K02739 20S proteasome subunit
beta 2 [EC:3.4.25.1]
Length=279
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 53/129 (41%), Positives = 73/129 (56%), Gaps = 18/129 (13%)
Query 15 GGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTGTTICGVIFQDG 74
GGF F N RN L + + P RKTGTTI G+I++DG
Sbjct 13 GGFSFENCPRNTFLQDQGPQLG-----------------LQPPKARKTGTTIAGIIYKDG 55
Query 75 VVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLATQVELHRLNTY 134
V+LGAD RAT+ +VADKNC K+H I+ NIY GAG AAD +++ L++ + +H ++T
Sbjct 56 VILGADRRATDDMVVADKNCAKIHYITDNIYCCGAGVAADAENVTQLLSSNLHIHAMST- 114
Query 135 AKQARVSMA 143
+Q RV A
Sbjct 115 GRQPRVCTA 123
> cel:C47B2.4 pbs-2; Proteasome Beta Subunit family member (pbs-2);
K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=277
Score = 95.1 bits (235), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 55/129 (42%), Positives = 73/129 (56%), Gaps = 22/129 (17%)
Query 15 GGFDFSNHCRNVSLLLRSAAASSSASSSTGVPSAAAAGSFRMPPLRKTGTTICGVIFQDG 74
G FDFSN RN ++ A P L TGTTI V F+ G
Sbjct 19 GAFDFSNCIRNQAMCKMGGKA---------------------PKLTSTGTTIVAVAFKGG 57
Query 75 VVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWLATQVELHRLNTY 134
+V+GAD+RAT G ++ADK+C K+HK++ +IYA GAGTAADLD + L+ + L LNT
Sbjct 58 LVMGADSRATAGNIIADKHCEKVHKLTESIYACGAGTAADLDQVTKMLSGNLRLLELNT- 116
Query 135 AKQARVSMA 143
++ARV A
Sbjct 117 GRKARVITA 125
> tgo:TGME49_023590 proteasome component PRE3 precursor, putative
(EC:3.4.25.1); K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=245
Score = 58.5 bits (140), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 54/100 (54%), Gaps = 5/100 (5%)
Query 38 SASSSTGVPSAAAAGSFRMPPLRKTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKL 97
+ASS G+P + + S R P TGTTI V F+ GVVLGADTR + G V ++ K+
Sbjct 20 AASSPAGLPVPSLSASGRGPTAVSTGTTIVAVSFKGGVVLGADTRTSAGSYVVNRAARKI 79
Query 98 HKISSNIYAAGAGTAADLDHMCDWLATQVELHRLNTYAKQ 137
++ I +G+AAD + TQ+ + YA++
Sbjct 80 SRVHERICVCRSGSAADTQAV-----TQIVKLYIQQYAQE 114
> hsa:5698 PSMB9, LMP2, MGC70470, PSMB6i, RING12, beta1i; proteasome
(prosome, macropain) subunit, beta type, 9 (large multifunctional
peptidase 2) (EC:3.4.25.1); K02741 20S proteasome
subunit beta 9 [EC:3.4.25.1]
Length=219
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTI V F GVV+G+D+R + G V ++ +KL + IY A +G+AAD + D
Sbjct 19 TGTTIMAVEFDGGVVMGSDSRVSAGEAVVNRVFDKLSPLHERIYCALSGSAADAQAVADM 78
Query 122 LATQVELHRLN 132
A Q+ELH +
Sbjct 79 AAYQLELHGIE 89
> dre:30665 psmb9a, lmp2, macropain, prosome, psmb9, wu:fa66f04,
wu:fj22d05, zgc:91842; proteasome (prosome, macropain) subunit,
beta type, 9a (EC:3.4.25.1); K02741 20S proteasome subunit
beta 9 [EC:3.4.25.1]
Length=218
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 61 KTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCD 120
KTGTTI V F GVV+G+D+R + G V ++ NKL + IY A +G+AAD + +
Sbjct 17 KTGTTIIAVTFDGGVVIGSDSRVSAGESVVNRVMNKLSPLHDKIYCALSGSAADAQTIAE 76
Query 121 WLATQVELHRLNT 133
+ Q+++H +
Sbjct 77 IVNYQLDVHSIEV 89
> mmu:19175 Psmb6, Lmp19, Mpnd; proteasome (prosome, macropain)
subunit, beta type 6 (EC:3.4.25.1); K02738 20S proteasome
subunit beta 1 [EC:3.4.25.1]
Length=238
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 42/71 (59%), Gaps = 0/71 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTI V F GVVLGAD+R T G +A++ +KL I +I+ +G+AAD + D
Sbjct 32 TGTTIMAVQFNGGVVLGADSRTTTGSYIANRVTDKLTPIHDHIFCCRSGSAADTQAVADA 91
Query 122 LATQVELHRLN 132
+ Q+ H +
Sbjct 92 VTYQLGFHSIE 102
> sce:YJL001W PRE3, CRL21; Beta 1 subunit of the 20S proteasome,
responsible for cleavage after acidic residues in peptides
(EC:3.4.25.1); K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=215
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 43/67 (64%), Gaps = 0/67 (0%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GT+I V F+DGV+LGAD+R T G +A++ +KL ++ I+ +G+AAD + D +
Sbjct 19 GTSIMAVTFKDGVILGADSRTTTGAYIANRVTDKLTRVHDKIWCCRSGSAADTQAIADIV 78
Query 123 ATQVELH 129
+EL+
Sbjct 79 QYHLELY 85
> xla:378667 psmb9, LMP2, psmb9-A; proteasome (prosome, macropain)
subunit, beta type, 9 (large multifunctional peptidase
2); K02741 20S proteasome subunit beta 9 [EC:3.4.25.1]
Length=215
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/74 (40%), Positives = 42/74 (56%), Gaps = 0/74 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTI V F GVVLG+D+R + G V ++ NKL + IY A +G+AAD + D
Sbjct 15 TGTTIIAVEFDGGVVLGSDSRVSAGDAVVNRVFNKLAPVHQRIYCALSGSAADAQAVADM 74
Query 122 LATQVELHRLNTYA 135
+E+H + A
Sbjct 75 AHYHMEVHSIEMEA 88
> hsa:5694 PSMB6, DELTA, LMPY, MGC5169, Y; proteasome (prosome,
macropain) subunit, beta type, 6 (EC:3.4.25.1); K02738 20S
proteasome subunit beta 1 [EC:3.4.25.1]
Length=239
Score = 55.8 bits (133), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTI V F GVVLGAD+R T G +A++ +KL I I+ +G+AAD + D
Sbjct 33 TGTTIMAVQFDGGVVLGADSRTTTGSYIANRVTDKLTPIHDRIFCCRSGSAADTQAVADA 92
Query 122 LATQVELHRLN 132
+ Q+ H +
Sbjct 93 VTYQLGFHSIE 103
> tpv:TP01_0874 proteasome subunit beta type 5; K02737 20S proteasome
subunit beta 5 [EC:3.4.25.1]
Length=266
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 47/83 (56%), Gaps = 3/83 (3%)
Query 61 KTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCD 120
K GTT G IF GV+L D+RA+ GP+V+ ++ +K+ +I+S + AG AAD +
Sbjct 53 KKGTTTLGFIFDHGVILAVDSRASMGPIVSTQSVSKVIEINSYLLGTMAGGAADCSYWER 112
Query 121 WLATQVELHRLNTYAKQARVSMA 143
LA LH L Q R+S+
Sbjct 113 HLAKLCRLHELRN---QCRISVG 132
> dre:30388 psmb6, y, zgc:109823; proteasome (prosome, macropain)
subunit, beta type, 6 (EC:3.4.25.1); K02738 20S proteasome
subunit beta 1 [EC:3.4.25.1]
Length=223
Score = 55.1 bits (131), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTI V F GVV+GAD+R T G +A++ +KL I I+ +G+AAD + D
Sbjct 22 TGTTIMAVEFDGGVVMGADSRTTTGAYIANRVTDKLTPIHDRIFCCRSGSAADTQAIADA 81
Query 122 LATQVELHRLN 132
+ Q+ H +
Sbjct 82 VTYQLGFHSIE 92
> ath:AT4G31300 PBA1; PBA1; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=234
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 44/69 (63%), Gaps = 0/69 (0%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTTI GV + GVVLGAD+R + G VA++ +K+ +++ N+Y +G+AAD + D++
Sbjct 12 GTTIIGVTYNGGVVLGADSRTSTGMYVANRASDKITQLTDNVYVCRSGSAADSQVVSDYV 71
Query 123 ATQVELHRL 131
+ H +
Sbjct 72 RYFLHQHTI 80
> xla:397716 proteasome subunit Y; K02738 20S proteasome subunit
beta 1 [EC:3.4.25.1]
Length=233
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTI V F GVV+GAD+R T G +A++ +KL + I+ +G+AAD + D
Sbjct 32 TGTTIMAVEFDGGVVIGADSRTTTGAYIANRVTDKLTPVHDRIFCCRSGSAADTQAIADA 91
Query 122 LATQVELHRLN 132
+ Q+ H +
Sbjct 92 VTYQLGFHSIE 102
> cpv:cgd2_860 Pre3p/proteasome regulatory subunit beta type 6,
NTN hydrolase fold ; K02738 20S proteasome subunit beta 1
[EC:3.4.25.1]
Length=248
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 45/73 (61%), Gaps = 0/73 (0%)
Query 61 KTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCD 120
+TGTTI + ++DG+VL AD R + GP++A + K+ +I+ ++ +G+AAD +
Sbjct 49 ETGTTIVALKYKDGLVLAADGRTSTGPIIAFRAARKITQITDKVFMCRSGSAADTQIISR 108
Query 121 WLATQVELHRLNT 133
++ V+ H L T
Sbjct 109 YVRRIVQDHELET 121
> xla:734745 hypothetical protein MGC130855; K02738 20S proteasome
subunit beta 1 [EC:3.4.25.1]
Length=247
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTTI V F GVV+GAD+R T G +A++ +KL + I+ +G+AAD + D
Sbjct 32 TGTTIMAVEFDGGVVIGADSRTTTGAYIANRVTDKLTPVHDRIFCCRSGSAADTQAIADA 91
Query 122 LATQVELHRLN 132
+ Q+ H +
Sbjct 92 VTYQLGFHSIE 102
> bbo:BBOV_IV009000 23.m05743; proteasome epsilon subunit; K02737
20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=268
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 48/83 (57%), Gaps = 3/83 (3%)
Query 61 KTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCD 120
K GTT G +F GV+L D+RA+ G ++A +N +K+ +I+S + AG AAD +
Sbjct 54 KKGTTTLGFVFNKGVILAVDSRASMGSIIATQNISKVIEINSYLLGTMAGGAADCTY--- 110
Query 121 WLATQVELHRLNTYAKQARVSMA 143
W +L RL+ Q R+++A
Sbjct 111 WERHVAKLCRLHELRNQERITVA 133
> tpv:TP02_0560 proteasome precursor; K02738 20S proteasome subunit
beta 1 [EC:3.4.25.1]
Length=279
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 42/69 (60%), Gaps = 4/69 (5%)
Query 61 KTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCD 120
K GTTI G+ F+DGVVL AD R + G +VA++ K+ +I NI+ +G+AAD
Sbjct 49 KLGTTIIGMKFEDGVVLVADGRTSSGQIVANRVARKITRILPNIFMLRSGSAAD----SQ 104
Query 121 WLATQVELH 129
L+T + H
Sbjct 105 TLSTIIRYH 113
> mmu:16912 Psmb9, Lmp-2, Lmp2; proteasome (prosome, macropain)
subunit, beta type 9 (large multifunctional peptidase 2) (EC:3.4.25.1);
K02741 20S proteasome subunit beta 9 [EC:3.4.25.1]
Length=219
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 46/78 (58%), Gaps = 1/78 (1%)
Query 48 AAAAGSFRMPPLRKTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAA 107
A AGSFR + TGTTI V F GVV+G+D+R + G V ++ +KL + +I+ A
Sbjct 6 APTAGSFRTEEVH-TGTTIMAVEFDGGVVVGSDSRVSAGTAVVNRVFDKLSPLHQHIFCA 64
Query 108 GAGTAADLDHMCDWLATQ 125
+G+AAD + D A Q
Sbjct 65 LSGSAADAQAIADMAAYQ 82
> bbo:BBOV_II005970 18.m06495; proteosome A; K02738 20S proteasome
subunit beta 1 [EC:3.4.25.1]
Length=266
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 37/52 (71%), Gaps = 0/52 (0%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAAD 114
GTTI +I++DG++L AD+R + GP+V ++ K+ +I N++ +G+AAD
Sbjct 37 GTTIIAMIYRDGILLAADSRTSSGPIVVNRVARKITRILPNVFMLRSGSAAD 88
> dre:64281 psmb9b; proteasome (prosome, macropain) subunit, beta
type, 9b (EC:3.4.25.1); K02741 20S proteasome subunit beta
9 [EC:3.4.25.1]
Length=227
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 42/71 (59%), Gaps = 0/71 (0%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTTI V F GVV+G+D+R + G V ++ NKL + IY A +G+AAD + + +
Sbjct 17 GTTIIAVEFDGGVVVGSDSRVSAGASVVNRVMNKLSPLHDKIYCALSGSAADAQTIAEIV 76
Query 123 ATQVELHRLNT 133
Q+++H +
Sbjct 77 NYQLDVHSIEV 87
> sce:YPR103W PRE2, DOA3, PRG1, SRR2; Beta 5 subunit of the 20S
proteasome, responsible for the chymotryptic activity of the
proteasome (EC:3.4.25.1); K02737 20S proteasome subunit beta
5 [EC:3.4.25.1]
Length=287
Score = 51.6 bits (122), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 43/81 (53%), Gaps = 3/81 (3%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTT FQ G+++ D+RAT G VA + K+ +I+ + AG AAD WL
Sbjct 75 GTTTLAFRFQGGIIVAVDSRATAGNWVASQTVKKVIEINPFLLGTMAGGAADCQFWETWL 134
Query 123 ATQVELHRLNTYAKQARVSMA 143
+Q LH L ++ R+S+A
Sbjct 135 GSQCRLHELR---EKERISVA 152
> cpv:cgd5_4210 Pre2p/proteasome subunit beta type 5; NTN hydrolase
fold ; K02737 20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=290
Score = 48.5 bits (114), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 43/81 (53%), Gaps = 3/81 (3%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTT G I+Q GV+L D+RA++G +A + K+ +I+ + AG AAD + W
Sbjct 79 GTTTLGFIYQGGVILAVDSRASQGSYIASQEVKKIIQINDFLLGTMAGGAADCSY---WE 135
Query 123 ATQVELHRLNTYAKQARVSMA 143
+L RL R+S+A
Sbjct 136 RVLSKLCRLYELRNGERISVA 156
> pfa:PF10_0111 20S proteasome beta subunit, putative; K02737
20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=271
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTT F+DG+++ D+RA+ G ++ +N K+ +I+ NI AG AAD + +L
Sbjct 60 GTTTLAFKFKDGIIVAVDSRASMGSFISSQNVEKIIEINKNILGTMAGGAADCLYWEKYL 119
Query 123 ATQVELHRL 131
++++ L
Sbjct 120 GKIIKIYEL 128
> pfa:PFI1545c proteasome precursor, putative (EC:3.4.25.1); K02738
20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=282
Score = 48.1 bits (113), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 35/51 (68%), Gaps = 0/51 (0%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAA 113
GTTI G+I+ +GV+L D+R + G +++K K+++I+ N+Y +G +A
Sbjct 30 GTTIIGIIYDNGVMLACDSRTSSGTFISNKCSRKINRINENLYVCRSGASA 80
> dre:64279 psmb11, MGC92422, macropain, prosome, zgc:92422; proteasome
(prosome, macropain) subunit, beta type, 11 (EC:3.4.25.1);
K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=217
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 39/72 (54%), Gaps = 1/72 (1%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGP-LVADKNCNKLHKISSNIYAAGAGTAADLDHMCD 120
TGTTI V F GV++G+D+RA+ G V+ K NKL ++ I+ AG+ AD +
Sbjct 15 TGTTILAVKFNGGVIIGSDSRASMGESYVSSKTINKLIQVHDRIFCCIAGSLADAQAVTK 74
Query 121 WLATQVELHRLN 132
Q+ H +
Sbjct 75 MAKFQLSFHSIQ 86
> cel:K08D12.1 pbs-1; Proteasome Beta Subunit family member (pbs-1);
K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=239
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 39/70 (55%), Gaps = 0/70 (0%)
Query 62 TGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDW 121
TGTT+ + + GVV+G D+R + G + + NK+ I+ N+ +G+AAD + D
Sbjct 22 TGTTLIAMEYNGGVVVGTDSRTSAGSFITSRATNKITPITDNMVVCRSGSAADTQAIADI 81
Query 122 LATQVELHRL 131
++++ +
Sbjct 82 AKYHIDVYTM 91
> tgo:TGME49_018920 proteasome subunit beta type 5, putative (EC:3.4.25.1);
K02737 20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=309
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 2/69 (2%)
Query 46 PSAAAAGSFRMPPLRKTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIY 105
P +A AGS M K GTT G +F+ G++L D+RA+ G ++ ++ K+ +IS I
Sbjct 81 PPSAQAGSPVMA--MKKGTTTLGFVFKGGIILAVDSRASMGTYISSQSVVKVIEISDIIL 138
Query 106 AAGAGTAAD 114
AG AAD
Sbjct 139 GTMAGGAAD 147
> dre:100331926 proteasome (prosome, macropain) subunit, beta
type, 5-like
Length=269
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 7/90 (7%)
Query 58 PLRKT----GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAA 113
P RK GTT FQ GV++ D+RAT G +A + K+ +I+ + AG AA
Sbjct 56 PERKIEFLHGTTTLAFKFQHGVIVAVDSRATAGAYIASQTVKKVIEINPYLLGTMAGGAA 115
Query 114 DLDHMCDWLATQVELHRLNTYAKQARVSMA 143
D LA Q ++ L + R+S+A
Sbjct 116 DCSFWERLLARQCRIYELRN---KERISVA 142
> dre:30387 psmb5, etID309919.2, fb59c07, si:zc14a17.13, wu:fb59c07,
x, zgc:86820; proteasome (prosome, macropain) subunit,
beta type, 5 (EC:3.4.25.1); K02737 20S proteasome subunit
beta 5 [EC:3.4.25.1]
Length=269
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 7/90 (7%)
Query 58 PLRKT----GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAA 113
P RK GTT FQ GV++ D+RAT G +A + K+ +I+ + AG AA
Sbjct 56 PERKIEFLHGTTTLAFKFQHGVIVAVDSRATAGAYIASQTVKKVIEINPYLLGTMAGGAA 115
Query 114 DLDHMCDWLATQVELHRLNTYAKQARVSMA 143
D LA Q ++ L + R+S+A
Sbjct 116 DCSFWERLLARQCRIYELRN---KERISVA 142
> xla:379720 MGC69086; low molecular mass protein-7 (LMP-7) homolog;
K02740 20S proteasome subunit beta 8 [EC:3.4.25.1]
Length=271
Score = 44.7 bits (104), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTT F+ GV++ D+RA+ G +A NK+ +I+ + +G+AAD H L
Sbjct 66 GTTTLAFKFKHGVIVAVDSRASAGSYIASLKANKVIEINPYLLGTMSGSAADCQHWERLL 125
Query 123 ATQVELHRLNTYAKQARVSMA 143
A + L++L +R+S++
Sbjct 126 AKECRLYQLRN---NSRISVS 143
> ath:AT3G26340 20S proteasome beta subunit E, putative; K02737
20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=273
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 45/81 (55%), Gaps = 3/81 (3%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTT IF++GV++ AD+RA+ G ++ ++ K+ +I+ + AG AAD L
Sbjct 57 GTTTLAFIFKEGVMVAADSRASMGGYISSQSVKKIIEINPYMLGTMAGGAADCQFWHRNL 116
Query 123 ATQVELHRLNTYAKQARVSMA 143
+ LH L A + R+S++
Sbjct 117 GIKCRLHEL---ANKRRISVS 134
> dre:30666 psmb8, lmp7, macropain, prosome; proteasome (prosome,
macropain) subunit, beta type, 8 (EC:3.4.25.1); K02740 20S
proteasome subunit beta 8 [EC:3.4.25.1]
Length=271
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTT F+ GV++ D+RA+ G + K NK+ +I+ + +G+AAD + L
Sbjct 67 GTTTLAFKFRHGVIVAVDSRASAGKYIDSKEANKVIEINPYLLGTMSGSAADCQYWERLL 126
Query 123 ATQVELHRLNTYAKQARVSMA 143
A + L++L + R+S++
Sbjct 127 AKECRLYKLRN---KQRISVS 144
> mmu:19173 Psmb5, MGC118075; proteasome (prosome, macropain)
subunit, beta type 5 (EC:3.4.25.1); K02737 20S proteasome subunit
beta 5 [EC:3.4.25.1]
Length=264
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/107 (32%), Positives = 51/107 (47%), Gaps = 10/107 (9%)
Query 37 SSASSSTGVPSAAAAGSFRMPPLRKTGTTICGVIFQDGVVLGADTRATEGPLVADKNCNK 96
S A+ S GVP R+ L GTT F GV++ AD+RAT G +A + K
Sbjct 40 SLAAPSWGVPEEP-----RIEMLH--GTTTLAFKFLHGVIVAADSRATAGAYIASQTVKK 92
Query 97 LHKISSNIYAAGAGTAADLDHMCDWLATQVELHRLNTYAKQARVSMA 143
+ +I+ + AG AAD LA Q ++ L + R+S+A
Sbjct 93 VIEINPYLLGTMAGGAADCSFWERLLARQCRIYELRN---KERISVA 136
> ath:AT1G13060 PBE1; PBE1; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02737 20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=274
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 63 GTTICGVIFQDGVVLGADTRATEGPLVADKNCNKLHKISSNIYAAGAGTAADLDHMCDWL 122
GTT IF+ GV++ AD+RA+ G ++ ++ K+ +I+ + AG AAD L
Sbjct 57 GTTTLAFIFKGGVMVAADSRASMGGYISSQSVKKIIEINPYMLGTMAGGAADCQFWHRNL 116
Query 123 ATQVELHRLNTYAKQARVSMA 143
+ LH L A + R+S++
Sbjct 117 GIKCRLHEL---ANKRRISVS 134
Lambda K H
0.318 0.128 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40