bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3651_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
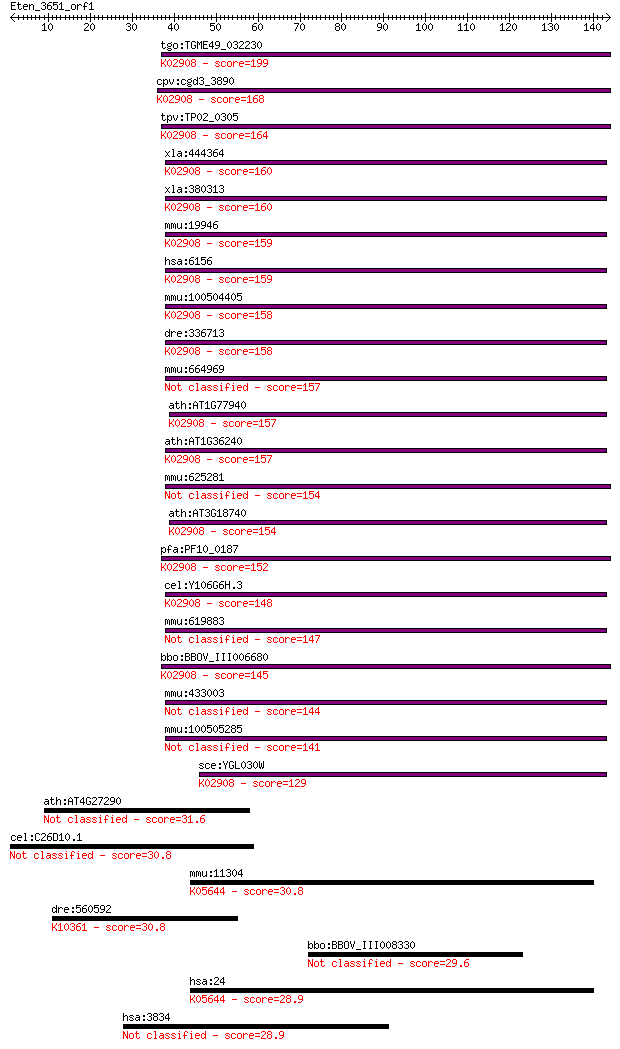
tgo:TGME49_032230 60S ribosomal protein L30, putative ; K02908... 199 3e-51
cpv:cgd3_3890 60S ribosomal protein L30, pelota RNA binding do... 168 5e-42
tpv:TP02_0305 60S ribosomal protein L30; K02908 large subunit ... 164 1e-40
xla:444364 MGC82844 protein; K02908 large subunit ribosomal pr... 160 2e-39
xla:380313 rpl30, MGC64253; ribosomal protein L30; K02908 larg... 160 2e-39
mmu:19946 Rpl30, MGC106425, MGC107559; ribosomal protein L30; ... 159 3e-39
hsa:6156 RPL30; ribosomal protein L30; K02908 large subunit ri... 159 3e-39
mmu:100504405 60S ribosomal protein L30-like; K02908 large sub... 158 5e-39
dre:336713 rpl30, fa93d05, wu:fa93d05, zgc:56640, zgc:77683; r... 158 5e-39
mmu:664969 Gm7429, EG664969; predicted pseudogene 7429 157 8e-39
ath:AT1G77940 60S ribosomal protein L30 (RPL30B); K02908 large... 157 9e-39
ath:AT1G36240 60S ribosomal protein L30 (RPL30A); K02908 large... 157 1e-38
mmu:625281 Gm6570, EG625281; predicted gene 6570 154 7e-38
ath:AT3G18740 60S ribosomal protein L30 (RPL30C); K02908 large... 154 9e-38
pfa:PF10_0187 60S ribosomal protein L30e, putative; K02908 lar... 152 4e-37
cel:Y106G6H.3 rpl-30; Ribosomal Protein, Large subunit family ... 148 5e-36
mmu:619883 Gm6109, EG619883; predicted gene 6109 147 2e-35
bbo:BBOV_III006680 17.m07591; ribosomal protein L7Ae containin... 145 5e-35
mmu:433003 Gm5481, EG433003; predicted gene 5481 144 1e-34
mmu:100505285 60S ribosomal protein L30-like 141 8e-34
sce:YGL030W RPL30; Rpl30p; K02908 large subunit ribosomal prot... 129 3e-30
ath:AT4G27290 ATP binding / protein kinase/ protein serine/thr... 31.6 0.94
cel:C26D10.1 ran-3; associated with RAN (nuclear import/export... 30.8 1.5
mmu:11304 Abca4, AW050280, Abc10, Abcr, D430003I15Rik, RmP; AT... 30.8 1.6
dre:560592 myo15aa, myo15l1; myosin XVAa; K10361 myosin XV 30.8 1.7
bbo:BBOV_III008330 17.m07728; hypothetical protein 29.6 3.1
hsa:24 ABCA4, ABC10, ABCR, ARMD2, CORD3, DKFZp781N1972, FFM, F... 28.9 5.6
hsa:3834 KIF25, KNSL3, MGC163361; kinesin family member 25 28.9
> tgo:TGME49_032230 60S ribosomal protein L30, putative ; K02908
large subunit ribosomal protein L30e
Length=108
Score = 199 bits (505), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 97/108 (89%), Positives = 103/108 (95%), Gaps = 1/108 (0%)
Query 37 MAKK-QKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIE 95
MAKK +K G+EGINSRLQLVMKSGKV LG KTA+H+LRTGKAKLIIISNNCPALRKSEIE
Sbjct 1 MAKKAKKSGSEGINSRLQLVMKSGKVNLGCKTAIHALRTGKAKLIIISNNCPALRKSEIE 60
Query 96 YYAMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSVE 143
YYAML+KCGVHHY GDNNELGTACGKLFRVSCL+VTDPGDSDIIRSVE
Sbjct 61 YYAMLSKCGVHHYHGDNNELGTACGKLFRVSCLAVTDPGDSDIIRSVE 108
> cpv:cgd3_3890 60S ribosomal protein L30, pelota RNA binding
domain containing protein ; K02908 large subunit ribosomal protein
L30e
Length=111
Score = 168 bits (426), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 79/110 (71%), Positives = 94/110 (85%), Gaps = 2/110 (1%)
Query 36 KMAKK--QKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSE 93
KMAKK K +EG+ SRLQLVMKSGKVCLG ++ V S+R G A+L+IISNNCP LR+SE
Sbjct 2 KMAKKGSSKGKSEGMTSRLQLVMKSGKVCLGYRSTVKSIRNGTARLVIISNNCPPLRRSE 61
Query 94 IEYYAMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSVE 143
IEYYAML+K GVHH+ G NN+LGT+CGKL+RVSC++VTDPGDSDIIRS E
Sbjct 62 IEYYAMLSKIGVHHFQGGNNDLGTSCGKLYRVSCMTVTDPGDSDIIRSYE 111
> tpv:TP02_0305 60S ribosomal protein L30; K02908 large subunit
ribosomal protein L30e
Length=108
Score = 164 bits (414), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 77/108 (71%), Positives = 89/108 (82%), Gaps = 1/108 (0%)
Query 37 MAKKQK-QGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIE 95
M KK K + E +N RLQLVMKSGKVCLG K+ +LR GKA L+I+SNNCP LR+SEIE
Sbjct 1 MGKKSKSKLVENVNHRLQLVMKSGKVCLGFKSTKATLRNGKALLVILSNNCPPLRRSEIE 60
Query 96 YYAMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSVE 143
YYAMLAKCGVHHY GDNN+LGTACGK FRV C+++ D GDSDI+RSVE
Sbjct 61 YYAMLAKCGVHHYTGDNNDLGTACGKHFRVGCMAILDAGDSDIVRSVE 108
> xla:444364 MGC82844 protein; K02908 large subunit ribosomal
protein L30e
Length=116
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 86/105 (81%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 AKKTKKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> xla:380313 rpl30, MGC64253; ribosomal protein L30; K02908 large
subunit ribosomal protein L30e
Length=116
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 86/105 (81%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 AKKTKKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> mmu:19946 Rpl30, MGC106425, MGC107559; ribosomal protein L30;
K02908 large subunit ribosomal protein L30e
Length=115
Score = 159 bits (402), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 86/105 (81%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 AKKTKKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> hsa:6156 RPL30; ribosomal protein L30; K02908 large subunit
ribosomal protein L30e
Length=115
Score = 159 bits (402), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 86/105 (81%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 AKKTKKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> mmu:100504405 60S ribosomal protein L30-like; K02908 large subunit
ribosomal protein L30e
Length=115
Score = 158 bits (400), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 74/105 (70%), Positives = 86/105 (81%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
+KK K+ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 SKKMKKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> dre:336713 rpl30, fa93d05, wu:fa93d05, zgc:56640, zgc:77683;
ribosomal protein L30; K02908 large subunit ribosomal protein
L30e
Length=117
Score = 158 bits (399), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 86/105 (81%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 AKKTKKSLESINSRLQLVMKSGKYVLGYKQSQKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> mmu:664969 Gm7429, EG664969; predicted pseudogene 7429
Length=115
Score = 157 bits (398), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 74/105 (70%), Positives = 85/105 (80%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQL MKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 AKKTKKSLESINSRLQLFMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> ath:AT1G77940 60S ribosomal protein L30 (RPL30B); K02908 large
subunit ribosomal protein L30e
Length=112
Score = 157 bits (397), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 75/104 (72%), Positives = 85/104 (81%), Gaps = 0/104 (0%)
Query 39 KKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYA 98
KK K+ EGINSRL LVMKSGK LG K+ + SLR K KLI+IS NCP LR+SEIEYYA
Sbjct 5 KKTKKSHEGINSRLALVMKSGKYTLGYKSVLKSLRGSKGKLILISTNCPPLRRSEIEYYA 64
Query 99 MLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
MLAK GVHHY G+N +LGTACGK FRVSCLS+ DPGDSDII+S+
Sbjct 65 MLAKVGVHHYNGNNVDLGTACGKYFRVSCLSIVDPGDSDIIKSI 108
> ath:AT1G36240 60S ribosomal protein L30 (RPL30A); K02908 large
subunit ribosomal protein L30e
Length=112
Score = 157 bits (397), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 87/105 (82%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ EGINSRL LVMKSGK LG K+ + SLR+ K KLI+IS+NCP LR+SEIEYY
Sbjct 4 AKKTKKSHEGINSRLALVMKSGKYTLGYKSVLKSLRSSKGKLILISSNCPPLRRSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY +N +LGTACGK FRVSCLS+ DPGDSDII+S+
Sbjct 64 AMLAKVGVHHYNRNNVDLGTACGKYFRVSCLSIVDPGDSDIIKSL 108
> mmu:625281 Gm6570, EG625281; predicted gene 6570
Length=115
Score = 154 bits (390), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 71/106 (66%), Positives = 87/106 (82%), Gaps = 0/106 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
A+K ++ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIE+Y
Sbjct 4 AEKTEKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEFY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSVE 143
AMLAK GV+HY G+N ELGTACGK +RV L++ DPGDSDIIRS++
Sbjct 64 AMLAKTGVYHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSMQ 109
> ath:AT3G18740 60S ribosomal protein L30 (RPL30C); K02908 large
subunit ribosomal protein L30e
Length=112
Score = 154 bits (389), Expect = 9e-38, Method: Compositional matrix adjust.
Identities = 73/104 (70%), Positives = 86/104 (82%), Gaps = 0/104 (0%)
Query 39 KKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYA 98
KK K+ EGINSRL LVMKSGK LG K+ + SLR+ K KLI+IS+NCP LR+SEIEYYA
Sbjct 5 KKAKKSHEGINSRLALVMKSGKYTLGYKSVLKSLRSSKGKLILISSNCPPLRRSEIEYYA 64
Query 99 MLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
MLAK GVH Y G+N +LGTACGK FRVSCLS+ DPGDSDII+++
Sbjct 65 MLAKVGVHRYNGNNVDLGTACGKYFRVSCLSIVDPGDSDIIKTL 108
> pfa:PF10_0187 60S ribosomal protein L30e, putative; K02908 large
subunit ribosomal protein L30e
Length=108
Score = 152 bits (384), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 71/108 (65%), Positives = 91/108 (84%), Gaps = 1/108 (0%)
Query 37 MAKKQKQGA-EGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIE 95
MAKK K+ A + IN++LQLVMKSGK G K+ + +LRTGK KL+I+S+NCP++++S IE
Sbjct 1 MAKKSKKSAGDNINAKLQLVMKSGKYQFGRKSCLKALRTGKGKLVIVSSNCPSIQRSVIE 60
Query 96 YYAMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSVE 143
YYAML+KCGVH Y GDNN+LGTACGKLFR+SCL +TD GDSDII++ E
Sbjct 61 YYAMLSKCGVHDYHGDNNDLGTACGKLFRISCLVITDVGDSDIIKTNE 108
> cel:Y106G6H.3 rpl-30; Ribosomal Protein, Large subunit family
member (rpl-30); K02908 large subunit ribosomal protein L30e
Length=113
Score = 148 bits (374), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 72/105 (68%), Positives = 83/105 (79%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
A K ++ AE INSRL +VMK+G+ LG K + SL GKAKL+II+NN P LRKSEIEYY
Sbjct 4 AAKPQKNAENINSRLSMVMKTGQYVLGYKQTLKSLLNGKAKLVIIANNTPPLRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACG+LFRV L+VTD GDSDII SV
Sbjct 64 AMLAKTGVHHYNGNNIELGTACGRLFRVCTLAVTDAGDSDIILSV 108
> mmu:619883 Gm6109, EG619883; predicted gene 6109
Length=115
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 69/105 (65%), Positives = 83/105 (79%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQLVM SGK L K + +R GKAKL+I++N+CP LRKSEIE+Y
Sbjct 4 AKKTKKSLESINSRLQLVMNSGKYVLSYKQTLKMVRQGKAKLVILANSCPDLRKSEIEFY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVH+Y G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHYYRGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> bbo:BBOV_III006680 17.m07591; ribosomal protein L7Ae containing
protein; K02908 large subunit ribosomal protein L30e
Length=108
Score = 145 bits (365), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 68/108 (62%), Positives = 84/108 (77%), Gaps = 1/108 (0%)
Query 37 MAKKQK-QGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIE 95
M KK K +G + +N +LQLVMKSGKVCLG K+ +LR GKA L+IISNNCP +R+SE+E
Sbjct 1 MVKKTKSKGLDSMNHKLQLVMKSGKVCLGFKSTKAALRNGKALLVIISNNCPPIRRSELE 60
Query 96 YYAMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSVE 143
+YAMLAKC VH + GDNNELGT CGK FRV ++V D GDSDI+R +
Sbjct 61 HYAMLAKCNVHMFAGDNNELGTVCGKYFRVGSMAVLDAGDSDILRGYD 108
> mmu:433003 Gm5481, EG433003; predicted gene 5481
Length=115
Score = 144 bits (363), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 68/105 (64%), Positives = 81/105 (77%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK ++ E INSRLQLVM SGK LG K + +R GKAKL+I++N+CPALRKSEIEYY
Sbjct 4 AKKTEKSLESINSRLQLVMNSGKYVLGYKQTLKMIRQGKAKLVILANSCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
MLAK G+H Y G N ELGTA GK +RV L++ DPGDSDIIRS+
Sbjct 64 VMLAKTGIHLYSGKNIELGTANGKHYRVWTLAIIDPGDSDIIRSM 108
> mmu:100505285 60S ribosomal protein L30-like
Length=115
Score = 141 bits (355), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 66/105 (62%), Positives = 79/105 (75%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
KK K+ E INSRLQ VMKSGK K + +R GKAKL+I++++CPA RKSEIEYY
Sbjct 4 TKKMKESLESINSRLQFVMKSGKYLREFKQTLKMVRQGKAKLVILADSCPAWRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N +LGTACGK +RV L+ DP DSDIIRS+
Sbjct 64 AMLAKTGVHHYRGNNMKLGTACGKYYRVCTLASIDPSDSDIIRSM 108
> sce:YGL030W RPL30; Rpl30p; K02908 large subunit ribosomal protein
L30e
Length=105
Score = 129 bits (324), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 59/97 (60%), Positives = 75/97 (77%), Gaps = 0/97 (0%)
Query 46 EGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYAMLAKCGV 105
E IN +L LV+KSGK LG K+ V SLR GK+KLIII+ N P LRKSE+EYYAML+K V
Sbjct 8 ESINQKLALVIKSGKYTLGYKSTVKSLRQGKSKLIIIAANTPVLRKSELEYYAMLSKTKV 67
Query 106 HHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
+++ G NNELGTA GKLFRV +S+ + GDSDI+ ++
Sbjct 68 YYFQGGNNELGTAVGKLFRVGVVSILEAGDSDILTTL 104
> ath:AT4G27290 ATP binding / protein kinase/ protein serine/threonine
kinase/ protein tyrosine kinase/ sugar binding
Length=783
Score = 31.6 bits (70), Expect = 0.94, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 9 APGKFGKGKFRPSAAAAAAAARAVCTPKMAKKQKQGAEGINSRLQLVMK 57
A K G+G F P A + V ++++ +QG E + ++L+ K
Sbjct 467 AGNKLGQGGFGPVYKGTLACGQEVAVKRLSRTSRQGVEEFKNEIKLIAK 515
> cel:C26D10.1 ran-3; associated with RAN (nuclear import/export)
function family member (ran-3)
Length=569
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Query 1 STSNGVSIAPGKFGKGKFRPSAAAAAAAARAVCTPKMAKKQKQGAEGINSRLQLVMKS 58
S + V P + G+ K PS + AAA + TP ++++G+E +N+ + M S
Sbjct 65 SIVDNVPKTPRRAGRPKKLPSTSTAAA--ESAVTPGKRGRKRKGSESVNTTINSTMNS 120
> mmu:11304 Abca4, AW050280, Abc10, Abcr, D430003I15Rik, RmP;
ATP-binding cassette, sub-family A (ABC1), member 4; K05644
ATP-binding cassette, subfamily A (ABC1), member 4
Length=2310
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 23/96 (23%), Positives = 41/96 (42%), Gaps = 2/96 (2%)
Query 44 GAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYAMLAKC 103
G + I + L +V +S +C H L + L + ++++E AML
Sbjct 990 GGKDIETNLDVVRQSLGMCPQHNILFHHLTVAEHILFYAQLKGRSWEEAQLEMEAMLEDT 1049
Query 104 GVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDII 139
G+HH N E G + R +++ GDS ++
Sbjct 1050 GLHH--KRNEEAQDLSGGMQRKLSVAIAFVGDSKVV 1083
> dre:560592 myo15aa, myo15l1; myosin XVAa; K10361 myosin XV
Length=4209
Score = 30.8 bits (68), Expect = 1.7, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 11 GKFGKGKFRPSAAAAAAAARAVCTPKMAKKQKQGAEGINSRLQL 54
GK GKGK A+ A A +AV PK + AE +LQL
Sbjct 93 GKRGKGKAALKGASKAVAVKAVGKPKRGQAADVKAEEKKPQLQL 136
> bbo:BBOV_III008330 17.m07728; hypothetical protein
Length=400
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 25/55 (45%), Gaps = 4/55 (7%)
Query 72 LRTGKAKLIIISNNCPALRKSEIEYYAMLAKCGVHHYPGD----NNELGTACGKL 122
L + K +++ N P L ++ + AKCG+H D N G CG++
Sbjct 23 LELLRQKALLVENFSPKLLDTQSHVIDIFAKCGMHISQDDVHIMRNRFGIPCGRV 77
> hsa:24 ABCA4, ABC10, ABCR, ARMD2, CORD3, DKFZp781N1972, FFM,
FLJ17534, RMP, RP19, STGD, STGD1; ATP-binding cassette, sub-family
A (ABC1), member 4; K05644 ATP-binding cassette, subfamily
A (ABC1), member 4
Length=2273
Score = 28.9 bits (63), Expect = 5.6, Method: Composition-based stats.
Identities = 22/96 (22%), Positives = 39/96 (40%), Gaps = 2/96 (2%)
Query 44 GAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYAMLAKC 103
G I + L V +S +C H L + L + ++++E AML
Sbjct 990 GGRDIETSLDAVRQSLGMCPQHNILFHHLTVAEHMLFYAQLKGKSQEEAQLEMEAMLEDT 1049
Query 104 GVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDII 139
G+HH N E G + R +++ GD+ ++
Sbjct 1050 GLHH--KRNEEAQDLSGGMQRKLSVAIAFVGDAKVV 1083
> hsa:3834 KIF25, KNSL3, MGC163361; kinesin family member 25
Length=332
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 28 AARAVCTPKMAKKQKQG-AEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNC 86
A++ P++ G AE + +RLQLV +G C+G G AKL++I
Sbjct 240 ASQGALAPQLVPGNPAGHAEQVQARLQLVDSAGSECVG----------GDAKLLVILCIS 289
Query 87 PALR 90
P+ R
Sbjct 290 PSQR 293
Lambda K H
0.317 0.131 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40