bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3638_orf2
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
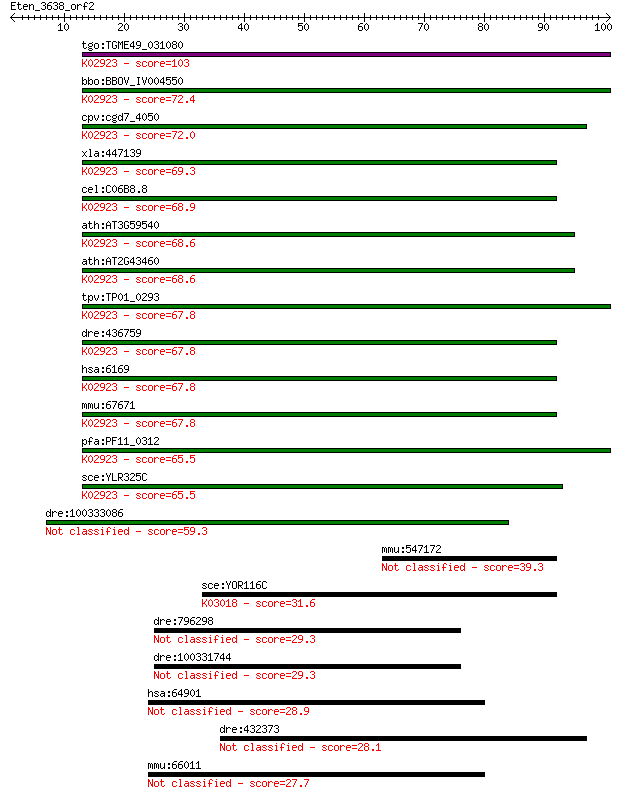
tgo:TGME49_031080 hypothetical protein ; K02923 large subunit ... 103 1e-22
bbo:BBOV_IV004550 23.m05864; ribosomal protein L38e; K02923 la... 72.4 4e-13
cpv:cgd7_4050 ribosomal protein L38 ; K02923 large subunit rib... 72.0 5e-13
xla:447139 rpl38, MGC85404; ribosomal protein L38; K02923 larg... 69.3 3e-12
cel:C06B8.8 rpl-38; Ribosomal Protein, Large subunit family me... 68.9 3e-12
ath:AT3G59540 60S ribosomal protein L38 (RPL38B); K02923 large... 68.6 5e-12
ath:AT2G43460 60S ribosomal protein L38 (RPL38A); K02923 large... 68.6 5e-12
tpv:TP01_0293 60S ribosomal protein L38; K02923 large subunit ... 67.8 8e-12
dre:436759 rpl38, MGC152655, wu:fb06b10, zgc:92860; ribosomal ... 67.8 8e-12
hsa:6169 RPL38; ribosomal protein L38; K02923 large subunit ri... 67.8 8e-12
mmu:67671 Rpl38, 0610025G13Rik, Ts; ribosomal protein L38; K02... 67.8 8e-12
pfa:PF11_0312 60S ribosomal protein L38e, putative; K02923 lar... 65.5 4e-11
sce:YLR325C RPL38; L38; K02923 large subunit ribosomal protein... 65.5 4e-11
dre:100333086 60S ribosomal protein L38-like 59.3 3e-09
mmu:547172 Gm6025, EG547172; predicted pseudogene 6025 39.3
sce:YOR116C RPO31, RPC1, RPC160; C160 (EC:2.7.7.6); K03018 DNA... 31.6 0.72
dre:796298 isotocin receptor-like 1 29.3
dre:100331744 Isotocin receptor-like 29.3 3.5
hsa:64901 RANBP17, FLJ32916; RAN binding protein 17 28.9
dre:432373 zgc:91897 28.1 6.3
mmu:66011 Ranbp17, 4932704E15Rik; RAN binding protein 17 27.7
> tgo:TGME49_031080 hypothetical protein ; K02923 large subunit
ribosomal protein L38e
Length=84
Score = 103 bits (257), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 50/88 (56%), Positives = 67/88 (76%), Gaps = 4/88 (4%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
MA+E+RD+R+FL TARR DA++V I R ++P+ A A+ +TKFKIRCS++LYT
Sbjct 1 MAKEIRDLRKFLLTARRPDAKRVTIVRQHKKPR----ATGGGASTVTKFKIRCSRYLYTF 56
Query 73 VVADKAKAQKIEGSLPPSLKQQIIPAKK 100
VV D+ KAQK+EGSLPPSL++ IP KK
Sbjct 57 VVEDREKAQKLEGSLPPSLEKVSIPGKK 84
> bbo:BBOV_IV004550 23.m05864; ribosomal protein L38e; K02923
large subunit ribosomal protein L38e
Length=79
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 53/88 (60%), Gaps = 10/88 (11%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M R L+D+R++L +R DA V +++ + V+TKFK+RCS+ LYTL
Sbjct 1 MPRALKDLRDYLAVLKRPDATAVTVYKK----------SGKGGVVLTKFKVRCSRFLYTL 50
Query 73 VVADKAKAQKIEGSLPPSLKQQIIPAKK 100
V + AKA KIE S+P LK+ +I +KK
Sbjct 51 TVPNHAKASKIEASIPSHLKKTVIASKK 78
> cpv:cgd7_4050 ribosomal protein L38 ; K02923 large subunit ribosomal
protein L38e
Length=82
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 36/84 (42%), Positives = 51/84 (60%), Gaps = 11/84 (13%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M ++ D+R+F+ T+RR DAR V + + TKFKIRCS++LYTL
Sbjct 4 MPIQITDLRDFINTSRRADARSVTV-----------VKVKKDNHITTKFKIRCSRYLYTL 52
Query 73 VVADKAKAQKIEGSLPPSLKQQII 96
V+ D KA+K+E SLPP+LK+ I
Sbjct 53 VMKDLEKAKKLESSLPPTLKKTTI 76
> xla:447139 rpl38, MGC85404; ribosomal protein L38; K02923 large
subunit ribosomal protein L38e
Length=70
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/79 (45%), Positives = 50/79 (63%), Gaps = 15/79 (18%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M R++ +I++FL TARRKDA+ V I ++ KFK+RCSK+LYTL
Sbjct 1 MPRKIEEIKDFLLTARRKDAKSVKIKKNKDN---------------VKFKVRCSKYLYTL 45
Query 73 VVADKAKAQKIEGSLPPSL 91
V+ DK KA+K++ SLPP L
Sbjct 46 VITDKEKAEKLKQSLPPGL 64
> cel:C06B8.8 rpl-38; Ribosomal Protein, Large subunit family
member (rpl-38); K02923 large subunit ribosomal protein L38e
Length=70
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 51/79 (64%), Gaps = 15/79 (18%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M +E+++I++FL ARRKDA+ V I ++ TKFK+RC+ +LYTL
Sbjct 1 MPKEIKEIKDFLVKARRKDAKSVKIKKNSNN---------------TKFKVRCASYLYTL 45
Query 73 VVADKAKAQKIEGSLPPSL 91
VVADK KA+K++ SLPP +
Sbjct 46 VVADKDKAEKLKQSLPPGI 64
> ath:AT3G59540 60S ribosomal protein L38 (RPL38B); K02923 large
subunit ribosomal protein L38e
Length=69
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 50/82 (60%), Gaps = 15/82 (18%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M +++ +I++FL TARRKDAR V I RS I KFK+RCS++LYTL
Sbjct 1 MPKQIHEIKDFLLTARRKDARSVKIKRS---------------KDIVKFKVRCSRYLYTL 45
Query 73 VVADKAKAQKIEGSLPPSLKQQ 94
V D+ KA K++ SLPP L Q
Sbjct 46 CVFDQEKADKLKQSLPPGLSVQ 67
> ath:AT2G43460 60S ribosomal protein L38 (RPL38A); K02923 large
subunit ribosomal protein L38e
Length=69
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 50/82 (60%), Gaps = 15/82 (18%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M +++ +I++FL TARRKDAR V I RS I KFK+RCS++LYTL
Sbjct 1 MPKQIHEIKDFLLTARRKDARSVKIKRS---------------KDIVKFKVRCSRYLYTL 45
Query 73 VVADKAKAQKIEGSLPPSLKQQ 94
V D+ KA K++ SLPP L Q
Sbjct 46 CVFDQEKADKLKQSLPPGLSVQ 67
> tpv:TP01_0293 60S ribosomal protein L38; K02923 large subunit
ribosomal protein L38e
Length=79
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 58/88 (65%), Gaps = 10/88 (11%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M +EL++++++L +R DAR VV+++ ++ KG + TKFK+RCS++LYT
Sbjct 1 MPKELKELKDYLSVLKRPDARSVVVYK--KKSKGGLLS--------TKFKVRCSRYLYTF 50
Query 73 VVADKAKAQKIEGSLPPSLKQQIIPAKK 100
V ++ KA K+E ++P L++++I KK
Sbjct 51 SVPNQVKAAKVEATIPSHLEKKVITNKK 78
> dre:436759 rpl38, MGC152655, wu:fb06b10, zgc:92860; ribosomal
protein L38; K02923 large subunit ribosomal protein L38e
Length=70
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 50/79 (63%), Gaps = 15/79 (18%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M R++ +I++FL TARRKDA+ V I ++ KFK+RCS++LYTL
Sbjct 1 MPRKIEEIKDFLLTARRKDAKSVKIKKNKDN---------------VKFKVRCSRYLYTL 45
Query 73 VVADKAKAQKIEGSLPPSL 91
V+ DK KA+K++ SLPP L
Sbjct 46 VITDKEKAEKLKQSLPPGL 64
> hsa:6169 RPL38; ribosomal protein L38; K02923 large subunit
ribosomal protein L38e
Length=70
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 50/79 (63%), Gaps = 15/79 (18%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M R++ +I++FL TARRKDA+ V I ++ KFK+RCS++LYTL
Sbjct 1 MPRKIEEIKDFLLTARRKDAKSVKIKKNKDN---------------VKFKVRCSRYLYTL 45
Query 73 VVADKAKAQKIEGSLPPSL 91
V+ DK KA+K++ SLPP L
Sbjct 46 VITDKEKAEKLKQSLPPGL 64
> mmu:67671 Rpl38, 0610025G13Rik, Ts; ribosomal protein L38; K02923
large subunit ribosomal protein L38e
Length=70
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 50/79 (63%), Gaps = 15/79 (18%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M R++ +I++FL TARRKDA+ V I ++ KFK+RCS++LYTL
Sbjct 1 MPRKIEEIKDFLLTARRKDAKSVKIKKNKDN---------------VKFKVRCSRYLYTL 45
Query 73 VVADKAKAQKIEGSLPPSL 91
V+ DK KA+K++ SLPP L
Sbjct 46 VITDKEKAEKLKQSLPPGL 64
> pfa:PF11_0312 60S ribosomal protein L38e, putative; K02923 large
subunit ribosomal protein L38e
Length=87
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 53/88 (60%), Gaps = 8/88 (9%)
Query 13 MARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTL 72
M +++ DIR+FL+ +R+ D V+I + + VITK K+R K+LYT+
Sbjct 1 MPKQITDIRKFLKISRKPDTTAVIIMKKK--------SKTKKNTVITKLKLRTKKYLYTM 52
Query 73 VVADKAKAQKIEGSLPPSLKQQIIPAKK 100
V AD+ KA++IE SL PSLK+ P KK
Sbjct 53 VFADRKKAERIENSLLPSLKRIYYPQKK 80
> sce:YLR325C RPL38; L38; K02923 large subunit ribosomal protein
L38e
Length=78
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 51/82 (62%), Gaps = 10/82 (12%)
Query 13 MARELRDIREFLQTARRKDARK--VVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLY 70
MARE+ DI++FL+ RR D + V I++ L + G PF TKFK+R S LY
Sbjct 1 MAREITDIKQFLELTRRADVKTATVKINKKLNKA-GKPFRQ-------TKFKVRGSSSLY 52
Query 71 TLVVADKAKAQKIEGSLPPSLK 92
TLV+ D KA+K+ SLPP+LK
Sbjct 53 TLVINDAGKAKKLIQSLPPTLK 74
> dre:100333086 60S ribosomal protein L38-like
Length=288
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 47/77 (61%), Gaps = 15/77 (19%)
Query 7 LSIQEKMARELRDIREFLQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCS 66
L+ ++ RE+ +I++FL TA+RKDA+ V I ++ KFK+RCS
Sbjct 11 LAGHGRLPREIEEIKDFLLTAKRKDAKSVKIKKNKDN---------------VKFKVRCS 55
Query 67 KHLYTLVVADKAKAQKI 83
+HLYTLV+ DK KA+K+
Sbjct 56 RHLYTLVITDKEKAEKL 72
> mmu:547172 Gm6025, EG547172; predicted pseudogene 6025
Length=76
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 22/29 (75%), Gaps = 0/29 (0%)
Query 63 IRCSKHLYTLVVADKAKAQKIEGSLPPSL 91
RC+++ YTLV+ DK KA+K++ SL P L
Sbjct 42 FRCNRYFYTLVITDKEKAEKLKQSLLPGL 70
> sce:YOR116C RPO31, RPC1, RPC160; C160 (EC:2.7.7.6); K03018 DNA-directed
RNA polymerase III subunit RPC1 [EC:2.7.7.6]
Length=1460
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 36/60 (60%), Gaps = 4/60 (6%)
Query 33 RKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCS-KHLYTLVVADKAKAQKIEGSLPPSL 91
++VV+ + +R KG F+A +AA ++ + ++ S + L+ L +K +A K G+L P +
Sbjct 2 KEVVVSETPKRIKGLEFSALSAADIVAQSEVEVSTRDLFDL---EKDRAPKANGALDPKM 58
> dre:796298 isotocin receptor-like 1
Length=382
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 25 QTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTLVVA 75
Q R K R + + R KGA + ++ +I+K KIR K + +V+A
Sbjct 233 QNFRLKTRRDQCLSLTPRPTKGAALSRVSSVKLISKAKIRTVKMTFVIVMA 283
> dre:100331744 Isotocin receptor-like
Length=382
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 25 QTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTLVVA 75
Q R K R + + R KGA + ++ +I+K KIR K + +V+A
Sbjct 233 QNFRLKTRRDQCLSLTPRPTKGAALSRVSSVKLISKAKIRTVKMTFVIVMA 283
> hsa:64901 RANBP17, FLJ32916; RAN binding protein 17
Length=1088
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 30/62 (48%), Gaps = 6/62 (9%)
Query 24 LQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITK------FKIRCSKHLYTLVVADK 77
L +R D R +++ +PK APF A VI K F+++ + ++ ++AD
Sbjct 77 LPVEQRMDIRNYILNYVASQPKLAPFVIQALIQVIAKITKLGWFEVQKDQFVFREIIADV 136
Query 78 AK 79
K
Sbjct 137 KK 138
> dre:432373 zgc:91897
Length=543
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 28/62 (45%), Gaps = 1/62 (1%)
Query 36 VIHRSLRRPKGAPFAAAAAAAVITKFKIRCSKHLYTLVVADKAK-AQKIEGSLPPSLKQQ 94
+I RSL F A V +F ++C + +V D K A+ IE S P SL+
Sbjct 291 LIQRSLEFAVSYVFTEAGEGVVFERFIVQCMNLIKMIVKNDAYKPAKNIEDSKPESLEAH 350
Query 95 II 96
I
Sbjct 351 RI 352
> mmu:66011 Ranbp17, 4932704E15Rik; RAN binding protein 17
Length=1088
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 30/62 (48%), Gaps = 6/62 (9%)
Query 24 LQTARRKDARKVVIHRSLRRPKGAPFAAAAAAAVITK------FKIRCSKHLYTLVVADK 77
L +R D R +++ +PK APF A VI K F+++ + ++ ++AD
Sbjct 77 LPIEQRIDIRNYILNYVASQPKLAPFVIQALIQVIAKLTKLGWFEVQKDEFVFREIIADV 136
Query 78 AK 79
K
Sbjct 137 KK 138
Lambda K H
0.323 0.133 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2041372988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40