bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3624_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
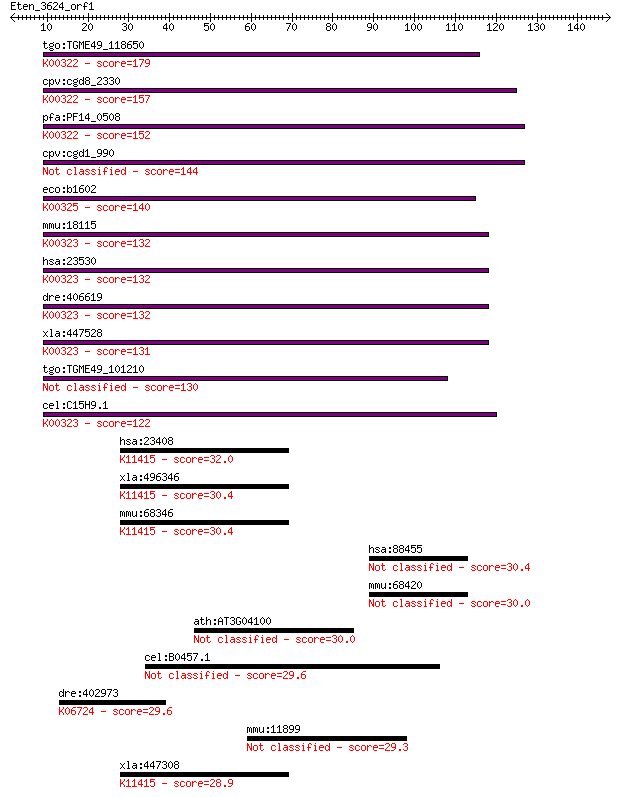
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 179 3e-45
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 157 8e-39
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 152 5e-37
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 144 1e-34
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 140 2e-33
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 132 3e-31
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 132 3e-31
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 132 5e-31
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 131 6e-31
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 130 2e-30
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 122 5e-28
hsa:23408 SIRT5, FLJ36950, SIR2L5; sirtuin 5; K11415 NAD-depen... 32.0 0.70
xla:496346 sirt5; sirtuin 5; K11415 NAD-dependent deacetylase ... 30.4 1.9
mmu:68346 Sirt5, 0610012J09Rik, 1500032M05Rik, AV001953; sirtu... 30.4 2.1
hsa:88455 ANKRD13A, ANKRD13, NY-REN-25; ankyrin repeat domain 13A 30.4 2.3
mmu:68420 Ankrd13a, 1100001D10Rik, ANKRD13, AU046136, MGC11850... 30.0 2.4
ath:AT3G04100 AGL57; AGL57; DNA binding / transcription factor 30.0 2.5
cel:B0457.1 lat-1; LATrophilin receptor family member (lat-1) 29.6 3.6
dre:402973 nrp1b; neuropilin 1b; K06724 neuropilin 1 29.6
mmu:11899 Astn1, Astn, GC14, mKIAA0289; astrotactin 1 29.3
xla:447308 MGC81733 protein; K11415 NAD-dependent deacetylase ... 28.9 5.8
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 83/107 (77%), Positives = 97/107 (90%), Gaps = 0/107 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIE 68
GHMNVLLAEA+V Y+ VKEMSEVN +M YDVV+VVGANDTVNPA+LEPG+KI GMPVIE
Sbjct 347 GHMNVLLAEANVSYRSVKEMSEVNKQMLEYDVVIVVGANDTVNPASLEPGTKIYGMPVIE 406
Query 69 AWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFA 115
WKA+RV VLKRS+A GYA+I+NPLF L+NTRMLFGNAK+TT+A+FA
Sbjct 407 VWKAKRVIVLKRSLAPGYAAIDNPLFFLDNTRMLFGNAKDTTTAIFA 453
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 157 bits (398), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 74/116 (63%), Positives = 90/116 (77%), Gaps = 2/116 (1%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIE 68
GHMNVLLAEADVPY IVKEM +VNP M S+DVVLV+GANDTVNP ALE SKI+GMPVIE
Sbjct 441 GHMNVLLAEADVPYSIVKEMEDVNPHMESFDVVLVIGANDTVNPLALEKDSKINGMPVIE 500
Query 69 AWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNAVCASP 124
W+A +V V KRS+ GYA+I+NPLF ++N M+FGNAK++ + N + SP
Sbjct 501 VWRASKVIVSKRSLGKGYAAIDNPLFFMKNVEMIFGNAKDSMINILQ--NIISISP 554
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 152 bits (383), Expect = 5e-37, Method: Composition-based stats.
Identities = 67/118 (56%), Positives = 93/118 (78%), Gaps = 0/118 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIE 68
GH+NVLLAEA++PY IVKEM+E+NP +S D+VLVVGAND VNP++L+P SKI GMPVIE
Sbjct 451 GHLNVLLAEANIPYNIVKEMNEINPIISEADIVLVVGANDIVNPSSLDPSSKIYGMPVIE 510
Query 69 AWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNAVCASPLP 126
WK+++V V KR++ GY++I+NPLF+ NT +LFG+AK+TT+ + +N + P
Sbjct 511 VWKSKQVIVFKRTLNTGYSAIDNPLFYFSNTFLLFGDAKHTTNQILTILNDYVNNKYP 568
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 144 bits (363), Expect = 1e-34, Method: Composition-based stats.
Identities = 73/118 (61%), Positives = 87/118 (73%), Gaps = 1/118 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIE 68
GHMNVLLAE+ VP +IVKEM VN M YD+VLVVGAND VNPAAL+P SKISGMPVI
Sbjct 404 GHMNVLLAESGVPSRIVKEMDAVNNCMHEYDLVLVVGANDIVNPAALDPQSKISGMPVIN 463
Query 69 AWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNAVCASPLP 126
WKA++V V KRS+A GYA IEN LF E TRML G+++ T V+ ++ CA +P
Sbjct 464 VWKAKQVVVSKRSLAYGYACIENELFTCERTRMLLGDSRETLQQVY-KILKGCAGFVP 520
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 140 bits (353), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 67/107 (62%), Positives = 80/107 (74%), Gaps = 1/107 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVI 67
GHMNVLLAEA VPY IV EM E+N + + D VLV+GANDTVNPAA + P S I+GMPV+
Sbjct 353 GHMNVLLAEAKVPYDIVLEMDEINDDFADTDTVLVIGANDTVNPAAQDDPKSPIAGMPVL 412
Query 68 EAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVF 114
E WKA+ V V KRSM GYA ++NPLF ENT MLFG+AK + A+
Sbjct 413 EVWKAQNVIVFKRSMNTGYAGVQNPLFFKENTHMLFGDAKASVDAIL 459
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 132 bits (333), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 63/110 (57%), Positives = 83/110 (75%), Gaps = 1/110 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVI 67
G +NVLLAEA VPY IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+
Sbjct 971 GQLNVLLAEAGVPYDIVLEMDEINSDFPDTDLVLVIGANDTVNSAAQEDPNSIIAGMPVL 1030
Query 68 EAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 117
E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1031 EVWKSKQVIVMKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKV 1080
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 132 bits (333), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 63/110 (57%), Positives = 83/110 (75%), Gaps = 1/110 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVI 67
G +NVLLAEA VPY IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+
Sbjct 971 GQLNVLLAEAGVPYDIVLEMDEINHDFPDTDLVLVIGANDTVNSAAQEDPNSIIAGMPVL 1030
Query 68 EAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 117
E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1031 EVWKSKQVIVMKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKV 1080
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 132 bits (331), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 62/110 (56%), Positives = 83/110 (75%), Gaps = 1/110 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVI 67
G +NVLLAEA VPY +V EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+
Sbjct 967 GQLNVLLAEAGVPYDVVLEMDEINEDFPETDLVLVIGANDTVNSAAQEDPNSIIAGMPVL 1026
Query 68 EAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 117
E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1027 EVWKSKQVVVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDALSAKV 1076
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 131 bits (330), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 62/110 (56%), Positives = 83/110 (75%), Gaps = 1/110 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVI 67
G +NVLLAEA VPY IV EM E+N + D+VLV+GANDTVN AA E P S I+GMPV+
Sbjct 971 GQLNVLLAEAGVPYDIVLEMDEINEDFPETDLVLVIGANDTVNSAAQEDPNSIIAGMPVL 1030
Query 68 EAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 117
E WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T ++ A+V
Sbjct 1031 EVWKSKQVIVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDSLQAKV 1080
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 61/100 (61%), Positives = 74/100 (74%), Gaps = 1/100 (1%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVI 67
GHMNVLLAEA+VPY IV M E+N + D V+++GANDTVNPAA PG I GMPV+
Sbjct 338 GHMNVLLAEANVPYDIVLSMDEINEDFPQTDTVIIIGANDTVNPAAQNAPGCPIYGMPVL 397
Query 68 EAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAK 107
E WKA++V VLKRSM GYA ++NPLF N+ ML G+AK
Sbjct 398 EVWKAKKVIVLKRSMRVGYAGVDNPLFFYPNSEMLLGDAK 437
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 122 bits (305), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 54/112 (48%), Positives = 80/112 (71%), Gaps = 1/112 (0%)
Query 9 GHMNVLLAEADVPYKIVKEMSEVNPEMSSYDVVLVVGANDTVNPAALE-PGSKISGMPVI 67
G +NVLLAEA VPY IV+EM E+N + DV LV+G+NDT+N AA + P S I+GMPV+
Sbjct 927 GQLNVLLAEAGVPYDIVEEMEEINEDFKETDVALVIGSNDTINSAAEDDPNSSIAGMPVL 986
Query 68 EAWKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNA 119
W +++V ++KR++ GYA+++NP+F ENT+ML G+AK + + V +
Sbjct 987 RVWNSKQVIIVKRTLGTGYAAVDNPVFFNENTQMLLGDAKKMSEKLLEEVKS 1038
> hsa:23408 SIRT5, FLJ36950, SIR2L5; sirtuin 5; K11415 NAD-dependent
deacetylase sirtuin 5 [EC:3.5.1.-]
Length=292
Score = 32.0 bits (71), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 28 MSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIE 68
+ EV+ E++ D+ LVVG + V PAA+ P G+PV E
Sbjct 214 LEEVDRELAHCDLCLVVGTSSVVYPAAMFAPQVAARGVPVAE 255
> xla:496346 sirt5; sirtuin 5; K11415 NAD-dependent deacetylase
sirtuin 5 [EC:3.5.1.-]
Length=309
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 1/42 (2%)
Query 28 MSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIE 68
+ EV E+ + D+ +VVG + V PAA+ P G+PV E
Sbjct 231 LGEVEKELETCDLCVVVGTSSVVYPAAMFAPQVAARGVPVAE 272
> mmu:68346 Sirt5, 0610012J09Rik, 1500032M05Rik, AV001953; sirtuin
5 (silent mating type information regulation 2 homolog)
5 (S. cerevisiae); K11415 NAD-dependent deacetylase sirtuin
5 [EC:3.5.1.-]
Length=310
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 28 MSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIE 68
+ EV+ E++ D+ LVVG + V PAA+ P G+PV E
Sbjct 232 LEEVDRELALCDLCLVVGTSSVVYPAAMFAPQVASRGVPVAE 273
> hsa:88455 ANKRD13A, ANKRD13, NY-REN-25; ankyrin repeat domain
13A
Length=590
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 89 IENPLFHLENTRMLFGNAKNTTSA 112
IE PLFH+ N R+ FGN ++A
Sbjct 411 IEIPLFHVLNARITFGNVNGCSTA 434
> mmu:68420 Ankrd13a, 1100001D10Rik, ANKRD13, AU046136, MGC118502;
ankyrin repeat domain 13a
Length=588
Score = 30.0 bits (66), Expect = 2.4, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 89 IENPLFHLENTRMLFGNAKNTTSA 112
IE PLFH+ N R+ FGN ++A
Sbjct 411 IEIPLFHVLNARITFGNVNGCSTA 434
> ath:AT3G04100 AGL57; AGL57; DNA binding / transcription factor
Length=207
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 46 ANDTVNPAALEPGSKISGMPVIEAWKARRVFVLKRSMAA 84
A+ NP+ EP + P++EA+K RR+ L + M A
Sbjct 78 ADRLKNPSRQEPLERDDTRPLVEAYKKRRLHDLVKKMEA 116
> cel:B0457.1 lat-1; LATrophilin receptor family member (lat-1)
Length=1012
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 32/72 (44%), Gaps = 0/72 (0%)
Query 34 EMSSYDVVLVVGANDTVNPAALEPGSKISGMPVIEAWKARRVFVLKRSMAAGYASIENPL 93
+M+ DV + + D S+I G +IE K RRV L + + + E+ +
Sbjct 428 QMTPSDVTVAIAGTDQTEVRKRRVVSRIVGASLIENGKERRVENLTQPVRITFYHKESSV 487
Query 94 FHLENTRMLFGN 105
HL N ++ N
Sbjct 488 RHLSNPTCVWWN 499
> dre:402973 nrp1b; neuropilin 1b; K06724 neuropilin 1
Length=959
Score = 29.6 bits (65), Expect = 4.0, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 1/26 (3%)
Query 13 VLLAEADVPYKIVKEMSEVNPEMSSY 38
VLL AD PY+++ E ++VNP S Y
Sbjct 814 VLLPSADTPYQVIIE-AQVNPTHSGY 838
> mmu:11899 Astn1, Astn, GC14, mKIAA0289; astrotactin 1
Length=1302
Score = 29.3 bits (64), Expect = 4.8, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 59 SKISGMPVIEAWKARRV-FVLKRSMAAGYASIENPLFHLE 97
S++SG PV++ WK R V + +K + AA + N L L+
Sbjct 806 SEVSGYPVLQHWKVRSVMYHIKLNQAAISQAFSNALHSLD 845
> xla:447308 MGC81733 protein; K11415 NAD-dependent deacetylase
sirtuin 5 [EC:3.5.1.-]
Length=309
Score = 28.9 bits (63), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 28 MSEVNPEMSSYDVVLVVGANDTVNPAAL-EPGSKISGMPVIE 68
+ EV E+ D+ +VVG + V PAA+ P G+PV E
Sbjct 231 LGEVEKELEMCDLCVVVGTSSVVYPAAMFAPQVAARGVPVAE 272
Lambda K H
0.317 0.131 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40