bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3618_orf2
Length=228
Score E
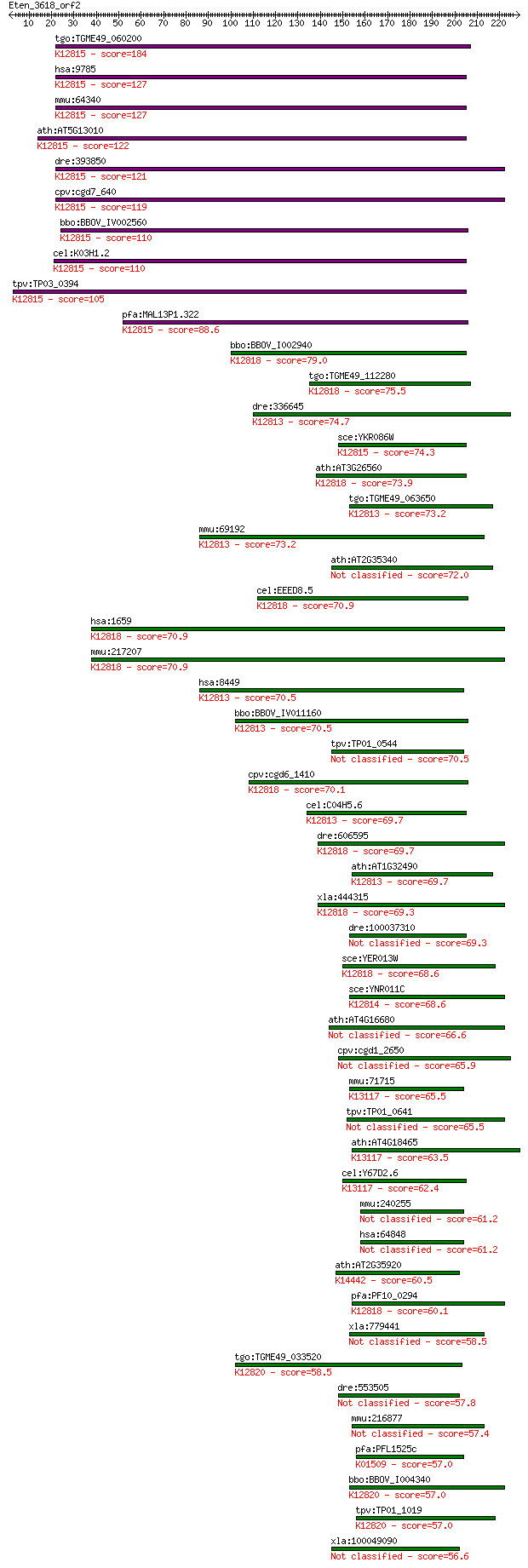
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_060200 ATP-dependent RNA helicase, putative (EC:3.6... 184 2e-46
hsa:9785 DHX38, DDX38, KIAA0224, PRP16, PRPF16; DEAH (Asp-Glu-... 127 2e-29
mmu:64340 Dhx38, 5730550P09Rik, AI325984, AW540902, Ddx38, Prp... 127 3e-29
ath:AT5G13010 EMB3011 (embryo defective 3011); ATP binding / R... 122 1e-27
dre:393850 dhx38, MGC63517, zgc:63517; DEAH (Asp-Glu-Ala-His) ... 121 2e-27
cpv:cgd7_640 Prp16p pre-mRNA splicing factor. HrpA family SFII... 119 1e-26
bbo:BBOV_IV002560 21.m03072; DEAH box RNA helicase (EC:3.6.1.-... 110 3e-24
cel:K03H1.2 mog-1; Masculinisation Of Germline family member (... 110 5e-24
tpv:TP03_0394 splicing factor; K12815 pre-mRNA-splicing factor... 105 2e-22
pfa:MAL13P1.322 splicing factor, putative; K12815 pre-mRNA-spl... 88.6 2e-17
bbo:BBOV_I002940 19.m02117; RNA helicase; K12818 ATP-dependent... 79.0 1e-14
tgo:TGME49_112280 ATP-dependent RNA helicase, putative (EC:3.4... 75.5 1e-13
dre:336645 dhx16, fa91b12, wu:fa91b12, zgc:55590; DEAH (Asp-Gl... 74.7 3e-13
sce:YKR086W PRP16, PRP23, RNA16; Prp16p (EC:3.6.1.-); K12815 p... 74.3 4e-13
ath:AT3G26560 ATP-dependent RNA helicase, putative; K12818 ATP... 73.9 4e-13
tgo:TGME49_063650 pre-mRNA splicing factor RNA helicase, putat... 73.2 7e-13
mmu:69192 Dhx16, 2410006N22Rik, DBP2, Ddx16, mKIAA0577; DEAH (... 73.2 8e-13
ath:AT2G35340 MEE29; MEE29 (maternal effect embryo arrest 29);... 72.0 2e-12
cel:EEED8.5 mog-5; Masculinisation Of Germline family member (... 70.9 3e-12
hsa:1659 DHX8, DDX8, HRH1, PRP22, PRPF22; DEAH (Asp-Glu-Ala-Hi... 70.9 3e-12
mmu:217207 Dhx8, Ddx8, KIAA4096, MGC31290, mDEAH6, mKIAA4096; ... 70.9 4e-12
hsa:8449 DHX16, DBP2, DDX16, PRO2014, PRP8, PRPF2, Prp2; DEAH ... 70.5 4e-12
bbo:BBOV_IV011160 23.m05966; RNA helicase (EC:3.6.1.-); K12813... 70.5 4e-12
tpv:TP01_0544 RNA helicase 70.5 5e-12
cpv:cgd6_1410 pre-mRNA splicing factor ATP-dependent RNA helic... 70.1 7e-12
cel:C04H5.6 mog-4; Masculinisation Of Germline family member (... 69.7 7e-12
dre:606595 im:7153552; K12818 ATP-dependent RNA helicase DHX8/... 69.7 8e-12
ath:AT1G32490 ESP3; ESP3 (ENHANCED SILENCING PHENOTYPE 3); ATP... 69.7 8e-12
xla:444315 MGC80994 protein; K12818 ATP-dependent RNA helicase... 69.3 1e-11
dre:100037310 zgc:158828 69.3 1e-11
sce:YER013W PRP22; DEAH-box RNA-dependent ATPase/ATP-dependent... 68.6 2e-11
sce:YNR011C PRP2, RNA2; Prp2p (EC:3.6.1.-); K12814 pre-mRNA-sp... 68.6 2e-11
ath:AT4G16680 RNA helicase, putative 66.6 6e-11
cpv:cgd1_2650 hypothetical protein 65.9 1e-10
mmu:71715 Dhx35, 1200009D07Rik, Ddx35; DEAH (Asp-Glu-Ala-His) ... 65.5 2e-10
tpv:TP01_0641 RNA helicase 65.5 2e-10
ath:AT4G18465 RNA helicase, putative; K13117 ATP-dependent RNA... 63.5 5e-10
cel:Y67D2.6 hypothetical protein; K13117 ATP-dependent RNA hel... 62.4 1e-09
mmu:240255 Ythdc2, 3010002F02Rik, BC037178; YTH domain contain... 61.2 3e-09
hsa:64848 YTHDC2; YTH domain containing 2 (EC:3.6.4.13) 61.2 3e-09
ath:AT2G35920 helicase domain-containing protein; K14442 ATP-d... 60.5 4e-09
pfa:PF10_0294 RNA helicase, putative; K12818 ATP-dependent RNA... 60.1 6e-09
xla:779441 dhx33, MGC154339; DEAH (Asp-Glu-Ala-His) box polype... 58.5 2e-08
tgo:TGME49_033520 ATP-dependent RNA helicase, putative (EC:3.4... 58.5 2e-08
dre:553505 hypothetical protein LOC553505 57.8 4e-08
mmu:216877 Dhx33, 3110057P17Rik, 9430096J02Rik, Ddx33; DEAH (A... 57.4 5e-08
pfa:PFL1525c pre-mRNA splicing factor RNA helicase, putative; ... 57.0 5e-08
bbo:BBOV_I004340 19.m02126; pre-mRNA splicing factor RNA helic... 57.0 5e-08
tpv:TP01_1019 ATP-dependent RNA helicase; K12820 pre-mRNA-spli... 57.0 6e-08
xla:100049090 dhx29; DEAH (Asp-Glu-Ala-His) box polypeptide 29... 56.6 6e-08
> tgo:TGME49_060200 ATP-dependent RNA helicase, putative (EC:3.6.1.15
2.7.1.127); K12815 pre-mRNA-splicing factor ATP-dependent
RNA helicase PRP16 [EC:3.6.4.13]
Length=1280
Score = 184 bits (468), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 97/195 (49%), Positives = 128/195 (65%), Gaps = 12/195 (6%)
Query 22 ETEDTEKEHVICRNVLPPFLKDYKPSDRIISVVKDDTSDLANLARKGSALLRYTRDATDR 81
E ED KEHVICRN+ PPFL + + +V+D TSD+ +ARKGSA+LR+T+D DR
Sbjct 324 EDEDEHKEHVICRNIRPPFLDGFDVEHSLPIMVQDVTSDMNVMARKGSAILRFTKDQEDR 383
Query 82 AAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAA-----AAAAAAAAGTRESY-----A 131
+AVRQRFWE+AGSTLG L+Q S A A+ A A + R+S+
Sbjct 384 SAVRQRFWELAGSTLGSLLQTSDEQAKAQLAQMQANNPWRVAGEDESEEDRQSFRQQNQY 443
Query 132 AAFLQAGKASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKT 191
AA L++ + + + ++++ EQR SLPV+ VR+EFL + REHQI++VVGETGSGKT
Sbjct 444 AAILRSSETE--ATSEFARTQSLAEQRRSLPVYAVRDEFLHIVREHQIVVVVGETGSGKT 501
Query 192 TQLTQYLLEDGYARP 206
TQLTQYL E GYA P
Sbjct 502 TQLTQYLFEAGYASP 516
> hsa:9785 DHX38, DDX38, KIAA0224, PRP16, PRPF16; DEAH (Asp-Glu-Ala-His)
box polypeptide 38 (EC:3.6.4.13); K12815 pre-mRNA-splicing
factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1227
Score = 127 bits (320), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 72/186 (38%), Positives = 101/186 (54%), Gaps = 11/186 (5%)
Query 22 ETEDTEKEHVICRNVLPPFLKD---YKPSDRIISVVKDDTSDLANLARKGSALLRYTRDA 78
E ++ K H++ N++PPFL + + VKD TSDLA +ARKGS +R R+
Sbjct 398 EEDNAAKVHLMVHNLVPPFLDGRIVFTKQPEPVIPVKDATSDLAIIARKGSQTVRKHREQ 457
Query 79 TDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAG 138
+R + + WE+AG+ LG +M EE A T + +A
Sbjct 458 KERKKAQHKHWELAGTKLGDIM----GVKKEEEPDKAVTEDGKVDYRTEQKFADHM---- 509
Query 139 KASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYL 198
K S + K++ EQR+ LP+F V+ E L + R++ I+IVVGETGSGKTTQLTQYL
Sbjct 510 KRKSEASSEFAKKKSILEQRQYLPIFAVQQELLTIIRDNSIVIVVGETGSGKTTQLTQYL 569
Query 199 LEDGYA 204
EDGY
Sbjct 570 HEDGYT 575
> mmu:64340 Dhx38, 5730550P09Rik, AI325984, AW540902, Ddx38, Prp16,
mKIAA0224; DEAH (Asp-Glu-Ala-His) box polypeptide 38 (EC:3.6.4.13);
K12815 pre-mRNA-splicing factor ATP-dependent
RNA helicase PRP16 [EC:3.6.4.13]
Length=1228
Score = 127 bits (319), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 72/186 (38%), Positives = 101/186 (54%), Gaps = 11/186 (5%)
Query 22 ETEDTEKEHVICRNVLPPFLKD---YKPSDRIISVVKDDTSDLANLARKGSALLRYTRDA 78
E ++ K H++ N++PPFL + + VKD TSDLA +ARKGS +R R+
Sbjct 399 EEDNAAKVHLMVHNLVPPFLDGRIVFTKQPEPVIPVKDATSDLAIIARKGSQTVRKHREQ 458
Query 79 TDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAG 138
+R + + WE+AG+ LG +M EE A T + +A
Sbjct 459 KERRKAQHKHWELAGTKLGDIM----GVKKEEEPDKAMTEDGKVDYRTEQKFADHM---- 510
Query 139 KASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYL 198
K S + K++ EQR+ LP+F V+ E L + R++ I+IVVGETGSGKTTQLTQYL
Sbjct 511 KEKSEASSEFAKKKSILEQRQYLPIFAVQQELLTIIRDNSIVIVVGETGSGKTTQLTQYL 570
Query 199 LEDGYA 204
EDGY
Sbjct 571 HEDGYT 576
> ath:AT5G13010 EMB3011 (embryo defective 3011); ATP binding /
RNA helicase/ helicase/ nucleic acid binding; K12815 pre-mRNA-splicing
factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1226
Score = 122 bits (306), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 74/194 (38%), Positives = 103/194 (53%), Gaps = 12/194 (6%)
Query 14 GAPLEGPEETEDTEKEHVICRNVLPPFLKD---YKPSDRIISVVKDDTSDLANLARKGSA 70
G ++ ++E+ K ++ + PPFL Y + VKD TSD+A ++RKGS
Sbjct 388 GTEVQTEFDSEEERKAILLVHDTKPPFLDGRVVYTKQAEPVMPVKDPTSDMAIISRKGSG 447
Query 71 LLRYTRDATDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESY 130
L++ R+ RQRFWE+AGS LG ++ +A E A A G +
Sbjct 448 LVKEIREKQSANKSRQRFWELAGSNLGNILGIEKSA-----EQIDADTAVVGDDGEVDFK 502
Query 131 AAAFLQAGKASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGK 190
A G + SKTM EQR+ LP+F VR+E L + RE+Q+++VVGETGSGK
Sbjct 503 GEAKFAQHMKKGEAVSEFAMSKTMAEQRQYLPIFSVRDELLQVIRENQVIVVVGETGSGK 562
Query 191 TTQLTQYLLEDGYA 204
TTQLTQ DGY
Sbjct 563 TTQLTQ----DGYT 572
> dre:393850 dhx38, MGC63517, zgc:63517; DEAH (Asp-Glu-Ala-His)
box polypeptide 38 (EC:3.6.4.13); K12815 pre-mRNA-splicing
factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1258
Score = 121 bits (303), Expect = 2e-27, Method: Composition-based stats.
Identities = 74/211 (35%), Positives = 108/211 (51%), Gaps = 17/211 (8%)
Query 22 ETEDTEKEHVICRNVLPPFLKD---YKPSDRIISVVKDDTSDLANLARKGSALLRYTRDA 78
E ++ + H++ N++PPFL + + VKD TSD+A ++RKGS L+R R+
Sbjct 427 EEDNAARVHLLVHNLVPPFLDGRIVFTKQPEPVIPVKDATSDMAIISRKGSQLVRRHREQ 486
Query 79 TDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAG 138
+R + + WE+AG+ LG +M ++E + A F
Sbjct 487 KERKKAQHKHWELAGTKLGDIM------GIQKKEDGGDSKAVGEDGKVDYRAEQKFADHM 540
Query 139 KASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYL 198
K S + KT+ EQR+ LP+F VR + L + R++ I+IVVGETGSGKTTQLTQYL
Sbjct 541 KEKSEASSDFAKKKTLLEQRQYLPIFAVRQQLLNIIRDNNIVIVVGETGSGKTTQLTQYL 600
Query 199 LEDGY--------ARPAAAAAAAAAAEAHPE 221
EDGY +P AA + A E
Sbjct 601 HEDGYTSYGMVGCTQPRRVAAMSVAKRVSEE 631
> cpv:cgd7_640 Prp16p pre-mRNA splicing factor. HrpA family SFII
helicase ; K12815 pre-mRNA-splicing factor ATP-dependent
RNA helicase PRP16 [EC:3.6.4.13]
Length=1042
Score = 119 bits (297), Expect = 1e-26, Method: Composition-based stats.
Identities = 75/226 (33%), Positives = 115/226 (50%), Gaps = 32/226 (14%)
Query 22 ETEDTEKEHVICRNVLPPFLK----------------DYKPSDRIISVVKDDTSDLANLA 65
+ ++ +K H+I P FLK D + + ++ +KD TSD+A LA
Sbjct 168 DEKNNKKIHIIVDRTYPKFLKSWELNTIKSQLESFKGDPETENDEVNTIKDKTSDIAKLA 227
Query 66 RKGSALLRYTRDATDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAA- 124
R GS++L+ R ++ +RQRFWE+ S +G L+ S + + EE S +
Sbjct 228 RTGSSILKTLRLKENKVKMRQRFWELQNSKIGALIS-SDKSHSIEENSIYSQDGPKEGKY 286
Query 125 -GTRESYAAAFLQAGKASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVV 183
+++S+ F ++ S + S + M R SLPV+KVR+ + L EH +++VV
Sbjct 287 DNSKQSFGDLFRKSKDIS-----ENDSRQRMLMTRRSLPVYKVRDSLIKLIGEHMVVVVV 341
Query 184 GETGSGKTTQLTQYLLEDGYAR--------PAAAAAAAAAAEAHPE 221
GETGSGKTTQLTQYL E GY++ P AA + A E
Sbjct 342 GETGSGKTTQLTQYLHEFGYSKRGIIGCTQPRRVAAVSVAQRVADE 387
> bbo:BBOV_IV002560 21.m03072; DEAH box RNA helicase (EC:3.6.1.-);
K12815 pre-mRNA-splicing factor ATP-dependent RNA helicase
PRP16 [EC:3.6.4.13]
Length=1016
Score = 110 bits (276), Expect = 3e-24, Method: Composition-based stats.
Identities = 74/208 (35%), Positives = 111/208 (53%), Gaps = 34/208 (16%)
Query 24 EDTEKEHVICRNVLPPFLKD-------------------------YKPSDRIISVVKDDT 58
++ K+ V+ R+V+PPF+ D K ++ ISVVKD T
Sbjct 174 KEEAKKIVLVRSVIPPFIYDGLSKETRDELDSSFAAGDNTFNHIYNKFLNQKISVVKDST 233
Query 59 SDLANLARKGSALLRYTRDATDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAA 118
SD+A +A+KGSA+LR ++ ++R A R RFW++ GS KL ++++S
Sbjct 234 SDIAQMAKKGSAILRKLKEESERNASRVRFWDLEGS---KLGSLLLLGDDSKDKSHDQPD 290
Query 119 AAAAAAGTRESYAAAFLQAG-KASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREH 177
+A +S + F+ S S S K ++E RE LPVFK R+E L+ +
Sbjct 291 NSA-----NDSKYSQFMDTDIPESELCERLSESKKKLRETREQLPVFKCRDELLSYIGQF 345
Query 178 QILIVVGETGSGKTTQLTQYLLEDGYAR 205
Q+++VVGETGSGKTTQL Q+L E GY +
Sbjct 346 QVMVVVGETGSGKTTQLAQFLYESGYYK 373
> cel:K03H1.2 mog-1; Masculinisation Of Germline family member
(mog-1); K12815 pre-mRNA-splicing factor ATP-dependent RNA
helicase PRP16 [EC:3.6.4.13]
Length=1131
Score = 110 bits (274), Expect = 5e-24, Method: Composition-based stats.
Identities = 71/188 (37%), Positives = 105/188 (55%), Gaps = 14/188 (7%)
Query 21 EETEDTEKEHVICRNVLPPFLKD----YKPSDRIISVVKDDTSDLANLARKGSALLRYTR 76
E+ D + ++ +N++PPFL K + II VV D T D+A A +GS +R R
Sbjct 307 EDETDENRVTILVQNIVPPFLDGRIVFTKQAQPIIPVV-DTTCDMAVSAARGSVAVRKRR 365
Query 77 DATDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQ 136
+ DR + + WE+AGS LG LM E++ A + +ES+ F
Sbjct 366 EVEDRKKAQDKHWELAGSKLGNLM------GVKEKKDETADPEDDDSGNYKESHQ--FAS 417
Query 137 AGKASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQ 196
K + A S + K++++QRE LPVF R + + + RE+ ++I+VGETGSGKTTQL Q
Sbjct 418 HMKDNEAVS-DFAMEKSIKQQREYLPVFACRQKMMNVIRENNVVIIVGETGSGKTTQLAQ 476
Query 197 YLLEDGYA 204
YLLEDG+
Sbjct 477 YLLEDGFG 484
> tpv:TP03_0394 splicing factor; K12815 pre-mRNA-splicing factor
ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1007
Score = 105 bits (261), Expect = 2e-22, Method: Composition-based stats.
Identities = 71/231 (30%), Positives = 118/231 (51%), Gaps = 36/231 (15%)
Query 3 LMQRGGAGPPGGAPLEGPEET---EDTEKEHVICRNVLPPFLKDY----KPSDRI----- 50
L Q GG A LE + +D K+ V+ RN++PPF+ + +D++
Sbjct 128 LRQGGGGSKFANAELEQLRASNVHKDETKKIVLVRNIIPPFIYEVYNCNNSTDQLTDELT 187
Query 51 -----------------ISVVKDDTSDLANLARKGSALLRYTRDATDRAAVRQRFWEVAG 93
+S VKD TSD+A +A+KGS +LR +D +R++ R RFW++
Sbjct 188 EAGDTFNDIYNKFLNQSVSTVKDPTSDIALMAKKGSQILRQMKDELERSSTRTRFWDLTN 247
Query 94 STLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKT 153
S +G L+ + + + +A+A G R Y + G+ +
Sbjct 248 SKIGNLIFKNNNRVGNS--MSNSGNNSASATGDRRGYMENEEKEGEEE-----LKKIKEH 300
Query 154 MQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA 204
++ R+SLPV++ ++E ++L ++ Q++I+VGETGSGKTTQL QYL E G+
Sbjct 301 LESVRKSLPVYQHKHEIISLIKQFQVIILVGETGSGKTTQLPQYLYESGFG 351
> pfa:MAL13P1.322 splicing factor, putative; K12815 pre-mRNA-splicing
factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1151
Score = 88.6 bits (218), Expect = 2e-17, Method: Composition-based stats.
Identities = 50/172 (29%), Positives = 88/172 (51%), Gaps = 19/172 (11%)
Query 52 SVVKDDTSDLANLARKGSALLRYTRDATDRAAVRQRFWEVAGSTLGKLMQGSAAAAAAEE 111
+VVKD+T D A+KGS L+Y + +++ R R+WE++ S LG+L++ +
Sbjct 353 TVVKDETCDFVKAAKKGSEFLKYFKSENEKSKARDRYWEISNSKLGELLKLYKNKKKNKN 412
Query 112 ESAAAAAAAAAAAGTRES------------------YAAAFLQAGKASGAGSGSSSSSKT 153
++ + + + E+ Y++ F + + +
Sbjct 413 DNTSKEDYDNISYNSSENNKEDDNGSDVFDYKKDKIYSSLF-NIENNNKDKKNTLKDKEE 471
Query 154 MQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR 205
+ + +ESLP++K ++E L + I+I+VGETGSGKTTQ+ QYL E+GY R
Sbjct 472 LLKLKESLPIYKSKHELLDAVYNNNIIIIVGETGSGKTTQIVQYLYEEGYHR 523
> bbo:BBOV_I002940 19.m02117; RNA helicase; K12818 ATP-dependent
RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1156
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 59/105 (56%), Gaps = 1/105 (0%)
Query 100 MQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTMQEQRE 159
+Q SA A + S + ++ A L + G ++ KT+QEQRE
Sbjct 439 IQRSAGAGSMTNNSTSQFMEELRRMNMKQRREGA-LHDKRDPGTRKDGHNAIKTIQEQRE 497
Query 160 SLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA 204
SLP+F +R+E L +E+ ILIVVGETGSGK+TQ+ QYL E GY
Sbjct 498 SLPIFALRDELLQAVQENDILIVVGETGSGKSTQIPQYLAESGYT 542
> tgo:TGME49_112280 ATP-dependent RNA helicase, putative (EC:3.4.22.44);
K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1206
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/72 (48%), Positives = 51/72 (70%), Gaps = 3/72 (4%)
Query 135 LQAGKASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQL 194
+ GK+ G S+K++ EQR+SLP++++R L +E+Q+LIV+GETGSGKTTQ+
Sbjct 528 MYIGKSVSFGQ---KSNKSIAEQRQSLPIYRLREPLLKAIKENQVLIVIGETGSGKTTQM 584
Query 195 TQYLLEDGYARP 206
TQYL E+G P
Sbjct 585 TQYLAEEGLVPP 596
> dre:336645 dhx16, fa91b12, wu:fa91b12, zgc:55590; DEAH (Asp-Glu-Ala-His)
box polypeptide 16 (EC:3.6.4.13); K12813 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX16 [EC:3.6.4.13]
Length=1054
Score = 74.7 bits (182), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/124 (36%), Positives = 62/124 (50%), Gaps = 17/124 (13%)
Query 110 EEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTMQEQRESLPVFKVRNE 169
EEE + A GT L ++ S + +++QE R SLP+F R +
Sbjct 369 EEEMITFVSTAITMKGT--------LSEKESEPELSQAEKQKQSIQEVRRSLPIFPYRED 420
Query 170 FLALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR---------PAAAAAAAAAAEAHP 220
LA +HQIL++ GETGSGKTTQ+ QYLLE+GY + P AA + AA
Sbjct 421 LLAAIGDHQILVIEGETGSGKTTQIPQYLLEEGYTKGGMKIGCTQPRRVAAMSVAARVAQ 480
Query 221 EAAA 224
E +
Sbjct 481 EMSV 484
> sce:YKR086W PRP16, PRP23, RNA16; Prp16p (EC:3.6.1.-); K12815
pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1071
Score = 74.3 bits (181), Expect = 4e-13, Method: Composition-based stats.
Identities = 30/57 (52%), Positives = 45/57 (78%), Gaps = 0/57 (0%)
Query 148 SSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA 204
+ S ++ +E LPVF+ R++ L+L RE+Q+++++GETGSGKTTQL QYL E+GYA
Sbjct 337 TPSKDDIKHTKEQLPVFRCRSQLLSLIRENQVVVIIGETGSGKTTQLAQYLYEEGYA 393
> ath:AT3G26560 ATP-dependent RNA helicase, putative; K12818 ATP-dependent
RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1168
Score = 73.9 bits (180), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 47/67 (70%), Gaps = 3/67 (4%)
Query 138 GKASGAGSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQY 197
GK G S S +QEQRESLP++K++ E + ++Q+L+V+GETGSGKTTQ+TQY
Sbjct 495 GKTPTFGQRSKLS---IQEQRESLPIYKLKKELIQAVHDNQVLVVIGETGSGKTTQVTQY 551
Query 198 LLEDGYA 204
L E GY
Sbjct 552 LAEAGYT 558
> tgo:TGME49_063650 pre-mRNA splicing factor RNA helicase, putative
(EC:3.4.22.44); K12813 pre-mRNA-splicing factor ATP-dependent
RNA helicase DHX16 [EC:3.6.4.13]
Length=1041
Score = 73.2 bits (178), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 47/72 (65%), Gaps = 8/72 (11%)
Query 153 TMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR------- 205
++QE+R+ LPV+ R EFL RE+ +LIVVGETGSGKTTQL QYL E GY +
Sbjct 391 SLQEERKMLPVYAFRTEFLRAVREYPVLIVVGETGSGKTTQLPQYLYEVGYGKAGKIGCT 450
Query 206 -PAAAAAAAAAA 216
P AA + AA
Sbjct 451 QPRRVAAMSVAA 462
> mmu:69192 Dhx16, 2410006N22Rik, DBP2, Ddx16, mKIAA0577; DEAH
(Asp-Glu-Ala-His) box polypeptide 16 (EC:3.6.4.13); K12813
pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [EC:3.6.4.13]
Length=1044
Score = 73.2 bits (178), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 53/130 (40%), Positives = 64/130 (49%), Gaps = 8/130 (6%)
Query 86 QRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAG- 144
QR WE A L G+ AAA E A T E AA LQ G +G
Sbjct 329 QRRWEEAQLGAASLKFGARDAAAQE----AKYQLVLEEDETIEFVRAAQLQ-GDEEPSGP 383
Query 145 --SGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDG 202
S + +++Q R SLPVF R E LA HQ+LI+ GETGSGKTTQ+ QYL E+G
Sbjct 384 PLSAQAQQKESIQAVRRSLPVFPFREELLAAIANHQVLIIEGETGSGKTTQIPQYLFEEG 443
Query 203 YARPAAAAAA 212
Y + A
Sbjct 444 YTKKGMKIAC 453
> ath:AT2G35340 MEE29; MEE29 (maternal effect embryo arrest 29);
ATP binding / ATP-dependent RNA helicase/ ATP-dependent helicase/
helicase/ nucleic acid binding
Length=1044
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/80 (43%), Positives = 50/80 (62%), Gaps = 8/80 (10%)
Query 145 SGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA 204
S S+ +QE R++LP++ R++ L ++HQ+LI+VGETGSGKTTQ+ QYL E GY
Sbjct 388 SAGKSAFHMLQEDRKALPIYTYRDQLLNAVKDHQVLIIVGETGSGKTTQIPQYLHEAGYT 447
Query 205 R--------PAAAAAAAAAA 216
+ P AA + AA
Sbjct 448 KLGKVGCTQPRRVAAMSVAA 467
> cel:EEED8.5 mog-5; Masculinisation Of Germline family member
(mog-5); K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1200
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 55/94 (58%), Gaps = 0/94 (0%)
Query 112 ESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTMQEQRESLPVFKVRNEFL 171
+S A +A + + +L+ A G + ++ +M EQRESLP+F ++ +
Sbjct 491 QSTAWSADESKDRNNKMKEMPEWLKHVTAGGKATYGRRTNLSMVEQRESLPIFALKKNLM 550
Query 172 ALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR 205
++QIL+VVGETGSGKTTQ+TQY +E G R
Sbjct 551 EAMIDNQILVVVGETGSGKTTQMTQYAIEAGLGR 584
> hsa:1659 DHX8, DDX8, HRH1, PRP22, PRPF22; DEAH (Asp-Glu-Ala-His)
box polypeptide 8 (EC:3.6.4.13); K12818 ATP-dependent RNA
helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1220
Score = 70.9 bits (172), Expect = 3e-12, Method: Composition-based stats.
Identities = 60/195 (30%), Positives = 88/195 (45%), Gaps = 18/195 (9%)
Query 38 PPFLKDYKPSDRIIS---VVKDDTSDLANLARKGSALLRYTRDATDRAAVRQRFWEVAGS 94
PPFL+ + +S +VK+ L+ A SAL + R+ + A R+ +
Sbjct 446 PPFLRGHTKQSMDMSPIKIVKNPDGSLSQAAMMQSALAKERREL--KQAQREAEMDSIPM 503
Query 95 TLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTM 154
L K A + +A E AF G + G + S +
Sbjct 504 GLNKHWVDPLPDAEGRQIAANMRGIGMMPNDIPEWKKHAF--GGNKASYGKKTQMS---I 558
Query 155 QEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY--------ARP 206
EQRESLP++K++ + + ++QILIV+GETGSGKTTQ+TQYL E GY +P
Sbjct 559 LEQRESLPIYKLKEQLVQAVHDNQILIVIGETGSGKTTQITQYLAEAGYTSRGKIGCTQP 618
Query 207 AAAAAAAAAAEAHPE 221
AA + A E
Sbjct 619 RRVAAMSVAKRVSEE 633
> mmu:217207 Dhx8, Ddx8, KIAA4096, MGC31290, mDEAH6, mKIAA4096;
DEAH (Asp-Glu-Ala-His) box polypeptide 8 (EC:3.6.4.13); K12818
ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1244
Score = 70.9 bits (172), Expect = 4e-12, Method: Composition-based stats.
Identities = 60/195 (30%), Positives = 88/195 (45%), Gaps = 18/195 (9%)
Query 38 PPFLKDYKPSDRIIS---VVKDDTSDLANLARKGSALLRYTRDATDRAAVRQRFWEVAGS 94
PPFL+ + +S +VK+ L+ A SAL + R+ + A R+ +
Sbjct 470 PPFLRGHTKQSMDMSPIKIVKNPDGSLSQAAMMQSALAKERREL--KQAQREAEMDSIPM 527
Query 95 TLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTM 154
L K A + +A E AF G + G + S +
Sbjct 528 GLNKHWVDPLPDAEGRQIAANMRGIGMMPNDIPEWKKHAF--GGNKASYGKKTQMS---I 582
Query 155 QEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY--------ARP 206
EQRESLP++K++ + + ++QILIV+GETGSGKTTQ+TQYL E GY +P
Sbjct 583 LEQRESLPIYKLKEQLVQAVHDNQILIVIGETGSGKTTQITQYLAEAGYTSRGKIGCTQP 642
Query 207 AAAAAAAAAAEAHPE 221
AA + A E
Sbjct 643 RRVAAMSVAKRVSEE 657
> hsa:8449 DHX16, DBP2, DDX16, PRO2014, PRP8, PRPF2, Prp2; DEAH
(Asp-Glu-Ala-His) box polypeptide 16 (EC:3.6.4.13); K12813
pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [EC:3.6.4.13]
Length=981
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 47/120 (39%), Positives = 60/120 (50%), Gaps = 6/120 (5%)
Query 86 QRFWEVAGSTLGKLMQGSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGS 145
QR WE A L G+ AA+ E + R A LQ + A
Sbjct 266 QRRWEEARLGAASLKFGARDAASQEPKYQLVLEEEETIEFVR----ATQLQGDEEPSAPP 321
Query 146 GSSSSSK--TMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY 203
S+ + + ++Q R SLPVF R E LA HQ+LI+ GETGSGKTTQ+ QYL E+GY
Sbjct 322 TSTQAQQKESIQAVRRSLPVFPFREELLAAIANHQVLIIEGETGSGKTTQIPQYLFEEGY 381
> bbo:BBOV_IV011160 23.m05966; RNA helicase (EC:3.6.1.-); K12813
pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16
[EC:3.6.4.13]
Length=931
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 38/104 (36%), Positives = 57/104 (54%), Gaps = 5/104 (4%)
Query 102 GSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTMQEQRESL 161
GS ++ + ES + + A+ R+++A L + + + E+R L
Sbjct 239 GSDSSDYEDSESDDELSDSTASTRKRKTFADRILAKQRRK-----QRKEHRKLLEERCRL 293
Query 162 PVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR 205
P++ R+E LA R H IL+VVGETGSGKTTQ+ QYL E GY +
Sbjct 294 PIYGYRHELLAAVRNHPILVVVGETGSGKTTQIPQYLYEVGYGK 337
> tpv:TP01_0544 RNA helicase
Length=910
Score = 70.5 bits (171), Expect = 5e-12, Method: Composition-based stats.
Identities = 30/59 (50%), Positives = 45/59 (76%), Gaps = 0/59 (0%)
Query 145 SGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY 203
+G +++++QE+R LP+FK+R+E + E+QILIV+GETGSGKTTQ+ QYL E +
Sbjct 133 AGEPRTNRSIQEERRGLPIFKLRDEIIREIIENQILIVIGETGSGKTTQIPQYLYESHF 191
> cpv:cgd6_1410 pre-mRNA splicing factor ATP-dependent RNA helicase
; K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1005
Score = 70.1 bits (170), Expect = 7e-12, Method: Composition-based stats.
Identities = 35/98 (35%), Positives = 60/98 (61%), Gaps = 4/98 (4%)
Query 108 AAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTMQEQRESLPVFKVR 167
A E A+A + T + +L GK+ G + ++S + EQR++LP++ +R
Sbjct 301 AGERTIASALRGIGMNSQTTPEWKRQYL--GKSLSFGKKNVTAS--ISEQRKNLPIYPMR 356
Query 168 NEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR 205
+ + R +Q+++V+GETGSGKTTQ+TQYL E+G+ +
Sbjct 357 DSLVDAIRNNQVIVVIGETGSGKTTQITQYLYEEGFCK 394
> cel:C04H5.6 mog-4; Masculinisation Of Germline family member
(mog-4); K12813 pre-mRNA-splicing factor ATP-dependent RNA
helicase DHX16 [EC:3.6.4.13]
Length=1008
Score = 69.7 bits (169), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 48/76 (63%), Gaps = 5/76 (6%)
Query 134 FLQAGKASGAG-----SGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGS 188
F+QA + G + + +++E R+SLPV+ R+ F+ +EHQ+LI+ GETGS
Sbjct 332 FIQALQMPGTNEEVVETEAEKKKMSIEETRKSLPVYAFRDAFIEAVKEHQVLIIEGETGS 391
Query 189 GKTTQLTQYLLEDGYA 204
GKTTQL QYL E G+
Sbjct 392 GKTTQLPQYLYEAGFC 407
> dre:606595 im:7153552; K12818 ATP-dependent RNA helicase DHX8/PRP22
[EC:3.6.4.13]
Length=1210
Score = 69.7 bits (169), Expect = 8e-12, Method: Composition-based stats.
Identities = 40/94 (42%), Positives = 56/94 (59%), Gaps = 11/94 (11%)
Query 139 KASGAGSGSSSSSKT---MQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLT 195
K + G+ +S KT + EQRESLP++K++ + + ++QILIV+GETGSGKTTQ+T
Sbjct 530 KHAFGGNKASYGKKTQLSILEQRESLPIYKLKEQLIQAVHDNQILIVIGETGSGKTTQIT 589
Query 196 QYLLEDGY--------ARPAAAAAAAAAAEAHPE 221
QYL E GY +P AA + A E
Sbjct 590 QYLAEAGYTTRGKIGCTQPRRVAAMSVAKRVSEE 623
> ath:AT1G32490 ESP3; ESP3 (ENHANCED SILENCING PHENOTYPE 3); ATP
binding / ATP-dependent RNA helicase/ ATP-dependent helicase/
helicase/ nucleic acid binding; K12813 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX16 [EC:3.6.4.13]
Length=1044
Score = 69.7 bits (169), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 33/71 (46%), Positives = 45/71 (63%), Gaps = 8/71 (11%)
Query 154 MQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR-------- 205
+QE R SLP++ R++ L EHQ+L++VG+TGSGKTTQ+ QYL E GY +
Sbjct 397 LQEVRRSLPIYTYRDQLLKAVEEHQVLVIVGDTGSGKTTQIPQYLHEAGYTKRGKVGCTQ 456
Query 206 PAAAAAAAAAA 216
P AA + AA
Sbjct 457 PRRVAAMSVAA 467
> xla:444315 MGC80994 protein; K12818 ATP-dependent RNA helicase
DHX8/PRP22 [EC:3.6.4.13]
Length=793
Score = 69.3 bits (168), Expect = 1e-11, Method: Composition-based stats.
Identities = 40/94 (42%), Positives = 55/94 (58%), Gaps = 11/94 (11%)
Query 139 KASGAGSGSSSSSKTMQ---EQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLT 195
K + G+ +S KT EQRESLP++K++ + + ++QILIV+GETGSGKTTQ+T
Sbjct 496 KHAFGGNKASYGKKTQMSILEQRESLPIYKLKEQLVQAVHDNQILIVIGETGSGKTTQIT 555
Query 196 QYLLEDGY--------ARPAAAAAAAAAAEAHPE 221
QYL E GY +P AA + A E
Sbjct 556 QYLAEAGYTTRGKIGCTQPRRVAAMSVAKRVSEE 589
> dre:100037310 zgc:158828
Length=250
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/52 (57%), Positives = 40/52 (76%), Gaps = 0/52 (0%)
Query 153 TMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA 204
T+++QR+ LPVFK RN L L Q +++VGETGSGK+TQ+ QYLLE G+A
Sbjct 44 TIEKQRQRLPVFKHRNNILYLVESFQTVVIVGETGSGKSTQIPQYLLEAGWA 95
> sce:YER013W PRP22; DEAH-box RNA-dependent ATPase/ATP-dependent
RNA helicase, associates with lariat intermediates before
the second catalytic step of splicing; mediates ATP-dependent
mRNA release from the spliceosome and unwinds RNA duplexes
(EC:3.6.1.-); K12818 ATP-dependent RNA helicase DHX8/PRP22
[EC:3.6.4.13]
Length=1145
Score = 68.6 bits (166), Expect = 2e-11, Method: Composition-based stats.
Identities = 34/79 (43%), Positives = 51/79 (64%), Gaps = 11/79 (13%)
Query 150 SSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA----- 204
+S + QR++LPV+ +R+E + R++Q L++VGETGSGKTTQ+TQYL E+G++
Sbjct 472 TSLPISAQRQTLPVYAMRSELIQAVRDNQFLVIVGETGSGKTTQITQYLDEEGFSNYGMI 531
Query 205 ------RPAAAAAAAAAAE 217
R AA + A AE
Sbjct 532 GCTQPRRVAAVSVAKRVAE 550
> sce:YNR011C PRP2, RNA2; Prp2p (EC:3.6.1.-); K12814 pre-mRNA-splicing
factor ATP-dependent RNA helicase-like protein PRP2
[EC:3.6.4.13]
Length=876
Score = 68.6 bits (166), Expect = 2e-11, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 50/79 (63%), Gaps = 10/79 (12%)
Query 153 TMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY--------- 203
T+QE R+ LPV + ++E L +++Q+LI++GETGSGKTTQL QYL+EDG+
Sbjct 215 TIQEARKLLPVHQYKDELLQEIKKNQVLIIMGETGSGKTTQLPQYLVEDGFTDQGKLQIA 274
Query 204 -ARPAAAAAAAAAAEAHPE 221
+P AA + AA E
Sbjct 275 ITQPRRVAATSVAARVADE 293
> ath:AT4G16680 RNA helicase, putative
Length=883
Score = 66.6 bits (161), Expect = 6e-11, Method: Composition-based stats.
Identities = 36/86 (41%), Positives = 50/86 (58%), Gaps = 8/86 (9%)
Query 144 GSGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY 203
G S ++++ +E RE LP+ R E L L E+Q+L++VGETGSGKTTQ+ QYL E GY
Sbjct 205 GCYSKTAAEKAREGREFLPIHGYREELLKLIEENQVLVIVGETGSGKTTQIPQYLQEAGY 264
Query 204 AR--------PAAAAAAAAAAEAHPE 221
+ P AA + A+ E
Sbjct 265 TKRGKIGCTQPRRVAAMSVASRVAQE 290
> cpv:cgd1_2650 hypothetical protein
Length=867
Score = 65.9 bits (159), Expect = 1e-10, Method: Composition-based stats.
Identities = 39/93 (41%), Positives = 49/93 (52%), Gaps = 16/93 (17%)
Query 148 SSSSKTMQEQ--------RESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLL 199
SS +K ++ Q R SLPV K + + + EH ILIVVGETGSGKTTQ+ QYL
Sbjct 179 SSRNKIIESQDIELINDVRNSLPVVKFKEQIIKSLEEHPILIVVGETGSGKTTQIPQYLF 238
Query 200 EDGY--------ARPAAAAAAAAAAEAHPEAAA 224
E GY +P AA + AA E +
Sbjct 239 EAGYYKNGIIACTQPRRVAAMSVAARVAKEMGS 271
> mmu:71715 Dhx35, 1200009D07Rik, Ddx35; DEAH (Asp-Glu-Ala-His)
box polypeptide 35 (EC:3.6.1.-); K13117 ATP-dependent RNA
helicase DDX35 [EC:3.6.4.13]
Length=679
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 26/51 (50%), Positives = 39/51 (76%), Gaps = 0/51 (0%)
Query 153 TMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY 203
++++QR+ LPVFK+RN L L +Q +++VGETG GK+TQ+ QYL E G+
Sbjct 46 SIEQQRQKLPVFKLRNHILYLVENYQTVVIVGETGCGKSTQIPQYLAEAGW 96
> tpv:TP01_0641 RNA helicase
Length=974
Score = 65.5 bits (158), Expect = 2e-10, Method: Composition-based stats.
Identities = 33/78 (42%), Positives = 47/78 (60%), Gaps = 8/78 (10%)
Query 152 KTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYAR------ 205
K + ++R+ LP++ R E L+ ++++ LIVVGETGSGKTTQ+ QYL E GY+R
Sbjct 300 KLILQERQKLPIYYYRTELLSAIKKYKTLIVVGETGSGKTTQIPQYLHEVGYSRAGVIGI 359
Query 206 --PAAAAAAAAAAEAHPE 221
P AA + A E
Sbjct 360 TQPRRVAAMSVATRVSKE 377
> ath:AT4G18465 RNA helicase, putative; K13117 ATP-dependent RNA
helicase DDX35 [EC:3.6.4.13]
Length=704
Score = 63.5 bits (153), Expect = 5e-10, Method: Composition-based stats.
Identities = 33/75 (44%), Positives = 43/75 (57%), Gaps = 0/75 (0%)
Query 154 MQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYARPAAAAAAA 213
+++QR+ LPV+K R E L L H I+VGETGSGKTTQ+ QYL E G+A A
Sbjct 41 IEKQRQRLPVYKYRTEILYLVENHATTIIVGETGSGKTTQIPQYLKEAGWAEGGRVIACT 100
Query 214 AAAEAHPEAAAAAAA 228
+A +A A
Sbjct 101 QPRRLAVQAVSARVA 115
> cel:Y67D2.6 hypothetical protein; K13117 ATP-dependent RNA helicase
DDX35 [EC:3.6.4.13]
Length=732
Score = 62.4 bits (150), Expect = 1e-09, Method: Composition-based stats.
Identities = 27/55 (49%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 150 SSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA 204
+S +Q+QR LP+FK R L +C ++ +I+VGETG GK+TQ+ Q+LLE G+A
Sbjct 66 ASLNIQQQRIRLPIFKNRGHILYMCERYRTIIIVGETGCGKSTQVPQFLLEAGWA 120
> mmu:240255 Ythdc2, 3010002F02Rik, BC037178; YTH domain containing
2 (EC:3.6.4.13)
Length=1445
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 39/46 (84%), Gaps = 0/46 (0%)
Query 158 RESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY 203
R+SLPVF+ + E + + +E++++++VGETGSGKTTQ+ Q+LL+D +
Sbjct 205 RQSLPVFEKQEEIVKIIKENKVVLIVGETGSGKTTQIPQFLLDDCF 250
> hsa:64848 YTHDC2; YTH domain containing 2 (EC:3.6.4.13)
Length=1430
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 23/46 (50%), Positives = 39/46 (84%), Gaps = 0/46 (0%)
Query 158 RESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY 203
R+SLPVF+ + E + + +E++++++VGETGSGKTTQ+ Q+LL+D +
Sbjct 190 RQSLPVFEKQEEIVKIIKENKVVLIVGETGSGKTTQIPQFLLDDCF 235
> ath:AT2G35920 helicase domain-containing protein; K14442 ATP-dependent
RNA helicase DHX36 [EC:3.6.4.13]
Length=995
Score = 60.5 bits (145), Expect = 4e-09, Method: Composition-based stats.
Identities = 27/55 (49%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 147 SSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLED 201
++ S K ++ RE LP FK++ EFL ++Q+L+V GETG GKTTQL Q++LE+
Sbjct 214 ATESVKALKAFREKLPAFKMKEEFLNSVSQNQVLVVSGETGCGKTTQLPQFILEE 268
> pfa:PF10_0294 RNA helicase, putative; K12818 ATP-dependent RNA
helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1290
Score = 60.1 bits (144), Expect = 6e-09, Method: Composition-based stats.
Identities = 29/76 (38%), Positives = 43/76 (56%), Gaps = 8/76 (10%)
Query 154 MQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY--------AR 205
+ EQR LP++ ++N+ + ++ +LIV+GETGSGKTTQ+ QYL E Y +
Sbjct 624 INEQRSKLPIYNLKNDLMKAIEKNNVLIVIGETGSGKTTQIPQYLHEANYTEKGIVGCTQ 683
Query 206 PAAAAAAAAAAEAHPE 221
P AA + A E
Sbjct 684 PRRVAAMSIAKRVSEE 699
> xla:779441 dhx33, MGC154339; DEAH (Asp-Glu-Ala-His) box polypeptide
33
Length=266
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 40/60 (66%), Gaps = 0/60 (0%)
Query 153 TMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYARPAAAAAA 212
T++EQR +LP+++ R++ +A R+ ++++GETGSGKTTQ+ QYL E R A
Sbjct 42 TLEEQRRNLPIYQARSQIIAQLRKLDSVVIIGETGSGKTTQIPQYLYEASIGRQGVIAVT 101
> tgo:TGME49_033520 ATP-dependent RNA helicase, putative (EC:3.4.22.44);
K12820 pre-mRNA-splicing factor ATP-dependent RNA
helicase DHX15/PRP43 [EC:3.6.4.13]
Length=801
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/101 (36%), Positives = 53/101 (52%), Gaps = 5/101 (4%)
Query 102 GSAAAAAAEEESAAAAAAAAAAAGTRESYAAAFLQAGKASGAGSGSSSSSKTMQEQRESL 161
G A AA + AA+ AAA Y + G G+ S + E R+ L
Sbjct 70 GLPPAQAALPGTVGAASVAAAPL-----YPNGEVPEGINPYTGAPYSQRYYKILEGRKKL 124
Query 162 PVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDG 202
P + + FL L + ++ +I+VGETGSGKTTQ+TQ+L+E G
Sbjct 125 PSWNAKKNFLKLVKRNRTVILVGETGSGKTTQMTQFLIEAG 165
> dre:553505 hypothetical protein LOC553505
Length=1077
Score = 57.8 bits (138), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 40/54 (74%), Gaps = 0/54 (0%)
Query 148 SSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLED 201
S ++ + ++R+ LPVF+ R + L R H++L++ GETGSGK+TQ+ Q++LE+
Sbjct 561 SPLARRLLDERKQLPVFQHREQVLEALRHHRVLVIAGETGSGKSTQIPQFILEE 614
> mmu:216877 Dhx33, 3110057P17Rik, 9430096J02Rik, Ddx33; DEAH
(Asp-Glu-Ala-His) box polypeptide 33 (EC:3.6.4.13)
Length=698
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/59 (45%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 154 MQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYARPAAAAAA 212
++ QR SLP+F+ R + LA R +++GETGSGKTTQ+ QYL E G +R A
Sbjct 58 LELQRRSLPIFRARGQLLAQLRNLDNAVLIGETGSGKTTQIPQYLYEGGISRQGIIAVT 116
> pfa:PFL1525c pre-mRNA splicing factor RNA helicase, putative;
K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=1168
Score = 57.0 bits (136), Expect = 5e-08, Method: Composition-based stats.
Identities = 25/48 (52%), Positives = 36/48 (75%), Gaps = 0/48 (0%)
Query 156 EQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGY 203
++R+ LP++ R + L + ++ILI+VGETGSGK+TQLTQYL E Y
Sbjct 433 DERKRLPIYSYRYDILKAIKNNKILILVGETGSGKSTQLTQYLYECKY 480
> bbo:BBOV_I004340 19.m02126; pre-mRNA splicing factor RNA helicase;
K12820 pre-mRNA-splicing factor ATP-dependent RNA helicase
DHX15/PRP43 [EC:3.6.4.13]
Length=703
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 44/77 (57%), Gaps = 8/77 (10%)
Query 153 TMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYL--------LEDGYA 204
T+ E+R LP + R F+ L R +Q++I+VGETGSGKTTQ+ Q++ L+
Sbjct 51 TILEKRRELPAWSARKNFVKLLRRNQVIILVGETGSGKTTQIPQFVVNSKLNQGLQVAVT 110
Query 205 RPAAAAAAAAAAEAHPE 221
+P AA + AA E
Sbjct 111 QPRRVAAMSVAARVADE 127
> tpv:TP01_1019 ATP-dependent RNA helicase; K12820 pre-mRNA-splicing
factor ATP-dependent RNA helicase DHX15/PRP43 [EC:3.6.4.13]
Length=729
Score = 57.0 bits (136), Expect = 6e-08, Method: Composition-based stats.
Identities = 30/73 (41%), Positives = 43/73 (58%), Gaps = 11/73 (15%)
Query 156 EQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLEDGYA----------- 204
E R+ LP + R F+ L + +Q+L++VGETGSGKTTQ+TQ+ L+ G +
Sbjct 75 EGRKKLPAWSARKNFVKLVKRNQVLVLVGETGSGKTTQMTQFALDAGLSGLKPIAITQPR 134
Query 205 RPAAAAAAAAAAE 217
R AA + A AE
Sbjct 135 RVAAMSVATRVAE 147
> xla:100049090 dhx29; DEAH (Asp-Glu-Ala-His) box polypeptide
29 (EC:3.6.4.13)
Length=1362
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 27/57 (47%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 145 SGSSSSSKTMQEQRESLPVFKVRNEFLALCREHQILIVVGETGSGKTTQLTQYLLED 201
S S K + RE LPVF N L + H++++V GETGSGK+TQ+ Q+LLED
Sbjct 550 SRDSMKYKRLLNDREQLPVFARGNFILETLKRHRVIVVAGETGSGKSTQVPQFLLED 606
Lambda K H
0.311 0.124 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7632817556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40