bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3599_orf2
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
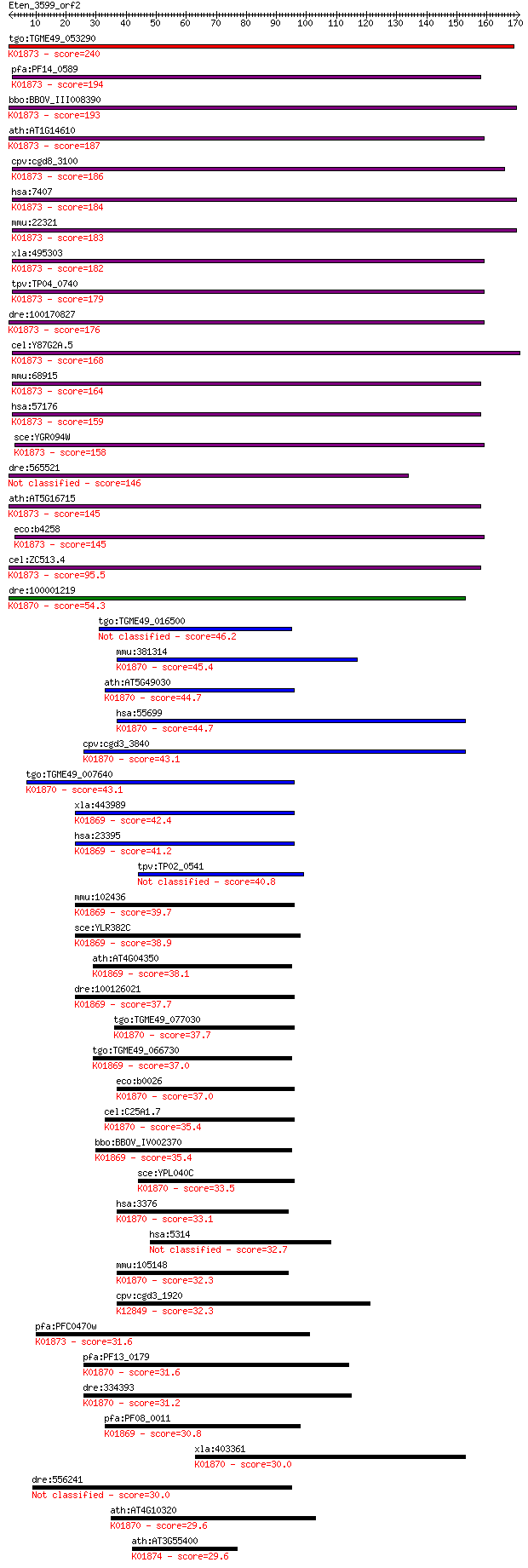
tgo:TGME49_053290 valyl-tRNA synthetase, putative (EC:6.1.1.9)... 240 1e-63
pfa:PF14_0589 valine-tRNA ligase, putative; K01873 valyl-tRNA ... 194 2e-49
bbo:BBOV_III008390 17.m07734; valyl-tRNA synthetase (EC:6.1.1.... 193 2e-49
ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRN... 187 1e-47
cpv:cgd8_3100 valyl-tRNA synthetase ; K01873 valyl-tRNA synthe... 186 2e-47
hsa:7407 VARS, G7A, VARS1, VARS2; valyl-tRNA synthetase (EC:6.... 184 9e-47
mmu:22321 Vars, Bat6, D17H6S56E, G7a, Vars2; valyl-tRNA synthe... 183 2e-46
xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9); K0... 182 6e-46
tpv:TP04_0740 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl... 179 3e-45
dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase ... 176 3e-44
cel:Y87G2A.5 vrs-2; Valyl tRNA Synthetase family member (vrs-2... 168 8e-42
mmu:68915 Vars2, 1190004I24Rik, Vars2l, mKIAA1885; valyl-tRNA ... 164 2e-40
hsa:57176 VARS2, MGC138259, MGC142165, VARS2L, VARSL; valyl-tR... 159 5e-39
sce:YGR094W VAS1; Mitochondrial and cytoplasmic valyl-tRNA syn... 158 1e-38
dre:565521 rCG46902-like 146 4e-35
ath:AT5G16715 EMB2247 (embryo defective 2247); ATP binding / a... 145 5e-35
eco:b4258 valS, ECK4251, JW4215; valyl-tRNA synthetase (EC:6.1... 145 7e-35
cel:ZC513.4 vrs-1; Valyl tRNA Synthetase family member (vrs-1)... 95.5 8e-20
dre:100001219 iars2, si:ch211-214p16.3, si:zc214p16.3; isoleuc... 54.3 2e-07
tgo:TGME49_016500 tRNA synthetase, putative (EC:6.1.1.9 6.1.1.4) 46.2 5e-05
mmu:381314 Iars2, 2010002H18Rik, C79125, MGC63429; isoleucine-... 45.4 8e-05
ath:AT5G49030 OVA2; OVA2 (ovule abortion 2); ATP binding / ami... 44.7 1e-04
hsa:55699 IARS2, FLJ10326, ILERS; isoleucyl-tRNA synthetase 2,... 44.7 1e-04
cpv:cgd3_3840 isoleucine-tRNA synthetase ; K01870 isoleucyl-tR... 43.1 4e-04
tgo:TGME49_007640 isoleucine-tRNA synthetase, putative (EC:6.1... 43.1 4e-04
xla:443989 lars2, MGC80460; leucyl-tRNA synthetase 2, mitochon... 42.4 8e-04
hsa:23395 LARS2, KIAA0028, LEURS, MGC26121; leucyl-tRNA synthe... 41.2 0.002
tpv:TP02_0541 hypothetical protein 40.8 0.002
mmu:102436 Lars2, AI035546, Kiaa0028, LEURS; leucyl-tRNA synth... 39.7 0.006
sce:YLR382C NAM2, MSL1; LeuRS; mitochondrial leucyl-tRNA synth... 38.9 0.008
ath:AT4G04350 EMB2369 (EMBRYO DEFECTIVE 2369); ATP binding / a... 38.1 0.013
dre:100126021 zgc:165522 (EC:6.1.1.4); K01869 leucyl-tRNA synt... 37.7 0.017
tgo:TGME49_077030 isoleucyl-tRNA synthetase, putative (EC:6.1.... 37.7 0.020
tgo:TGME49_066730 leucyl-tRNA synthetase, putative (EC:6.1.1.4... 37.0 0.030
eco:b0026 ileS, ECK0027, ilvS, JW0024; isoleucyl-tRNA syntheta... 37.0 0.033
cel:C25A1.7 irs-2; Isoleucyl tRNA Synthetase family member (ir... 35.4 0.10
bbo:BBOV_IV002370 21.m02840; leucyl-tRNA synthetase (EC:6.1.1.... 35.4 0.10
sce:YPL040C ISM1; Ism1p (EC:6.1.1.5); K01870 isoleucyl-tRNA sy... 33.5 0.35
hsa:3376 IARS, FLJ20736, IARS1, ILERS, ILRS, IRS, PRO0785; iso... 33.1 0.51
hsa:5314 PKHD1, ARPKD, DKFZp686C01112, FCYT, FLJ46150, TIGM1; ... 32.7 0.60
mmu:105148 Iars, 2510016L12Rik, AI327140, AU044614, E430001P04... 32.3 0.71
cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family m... 32.3 0.83
pfa:PFC0470w valine-tRNA ligase, putative (EC:6.1.1.9); K01873... 31.6 1.3
pfa:PF13_0179 isoleucine-tRNA ligase, putative (EC:6.1.1.5); K... 31.6 1.3
dre:334393 iars, fi46h05, wu:fi46h05, zgc:63790; isoleucyl-tRN... 31.2 1.6
pfa:PF08_0011 leucine-tRNA ligase; K01869 leucyl-tRNA syntheta... 30.8 2.5
xla:403361 iars, MGC68929; isoleucyl-tRNA synthetase; K01870 i... 30.0 3.7
dre:556241 very large inducible GTPase-1-like 30.0 3.7
ath:AT4G10320 isoleucyl-tRNA synthetase, putative / isoleucine... 29.6 5.0
ath:AT3G55400 OVA1; OVA1 (OVULE ABORTION 1); ATP binding / ami... 29.6 5.0
> tgo:TGME49_053290 valyl-tRNA synthetase, putative (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1042
Score = 240 bits (613), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 115/170 (67%), Positives = 138/170 (81%), Gaps = 2/170 (1%)
Query 1 LKEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVV 60
LKE SRH+ GR+ F+ KVW WK +YG+ I DQLRRVGSSVSW HF FTLD KLS AVV
Sbjct 119 LKEGAPSRHELGREAFLRKVWDWKRRYGDTICDQLRRVGSSVSWPHFSFTLDEKLSRAVV 178
Query 61 EAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLW 120
EAFVR+++ GLIYREERLVSWS LKTA+SD+EVD E I++P+ V IPGF++PVEVG+LW
Sbjct 179 EAFVRMYDAGLIYREERLVSWSPYLKTAISDVEVDVEEIDKPKKVTIPGFEYPVEVGYLW 238
Query 121 HFVYKVEGGGEIEVATTRVETMLGDVAVAVHPSDPRYQ--AGPSSLHFYF 168
HF Y+VEGGG +EVATTR+ETMLGDVAVAV+P+D RY G +H +F
Sbjct 239 HFSYEVEGGGRLEVATTRIETMLGDVAVAVNPTDERYSNLVGKRLIHPFF 288
> pfa:PF14_0589 valine-tRNA ligase, putative; K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1090
Score = 194 bits (492), Expect = 2e-49, Method: Composition-based stats.
Identities = 87/157 (55%), Positives = 121/157 (77%), Gaps = 1/157 (0%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
K+ENK R D+GR++FV+K+++WK+ +GN+I +Q++R+G+SV W FT++ LS AV E
Sbjct 144 KKENKIRQDYGREEFVKKIYEWKDLHGNKINNQMKRIGASVDWSREYFTMNENLSNAVKE 203
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AF++ + GLIYR+ RLV+W LKTALSDIEV+ E I++P ++IP FD VEVG L+
Sbjct 204 AFIKFYESGLIYRDNRLVAWCPHLKTALSDIEVNLEEIKKPTKIKIPSFDHLVEVGVLYK 263
Query 122 FVYKVEGGGE-IEVATTRVETMLGDVAVAVHPSDPRY 157
F Y+++ E IE+ATTR+ETMLGDVAVAVHP D RY
Sbjct 264 FFYQIKDSEEKIEIATTRIETMLGDVAVAVHPKDKRY 300
> bbo:BBOV_III008390 17.m07734; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=972
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 94/174 (54%), Positives = 127/174 (72%), Gaps = 5/174 (2%)
Query 1 LKEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVV 60
++ ENK+RHDF R+DF+++V+QW +QYGN I DQL R+G+S+ W FT+D+ S AV+
Sbjct 107 MQTENKTRHDFSREDFIKRVFQWNDQYGNNIKDQLGRMGASLDWTRDAFTMDAPRSKAVI 166
Query 61 EAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLW 120
EAFVR++N G IYR RLVSW L TALSDIEVDT + P T+++PG++ VEVG+LW
Sbjct 167 EAFVRMYNDGFIYRATRLVSWCPHLSTALSDIEVDTFDVTSPVTIKVPGYEKSVEVGWLW 226
Query 121 HFVYKVEGGGEIE---VATTRVETMLGDVAVAVHPSDPRYQ--AGPSSLHFYFS 169
F Y+V+G ++ VATTR+ETMLGDVAVAV P+D RY+ G +H + S
Sbjct 227 IFKYEVKGSSPVQYISVATTRIETMLGDVAVAVLPNDNRYRDYVGKELVHPFVS 280
> ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / valine-tRNA ligase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1108
Score = 187 bits (476), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 85/159 (53%), Positives = 118/159 (74%), Gaps = 1/159 (0%)
Query 1 LKEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVV 60
+++ +RHD GR++FV++VW+WK QYG I QLRR+G+S+ W CFT+D + S AV
Sbjct 230 MRDRGMTRHDVGREEFVKEVWKWKNQYGGTILTQLRRLGASLDWSRECFTMDEQRSKAVT 289
Query 61 EAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLW 120
EAFVRL+ GLIYR+ RLV+W L+TA+SD+EV+ I++ +++PG++ PVE G L
Sbjct 290 EAFVRLYKEGLIYRDIRLVNWDCILRTAISDVEVEYIDIKEKTLLKVPGYEKPVEFGLLT 349
Query 121 HFVYKVEGG-GEIEVATTRVETMLGDVAVAVHPSDPRYQ 158
F Y +EGG GE+ VATTRVETMLGD A+A+HP D RY+
Sbjct 350 SFAYPLEGGLGEVIVATTRVETMLGDTAIAIHPDDARYK 388
> cpv:cgd8_3100 valyl-tRNA synthetase ; K01873 valyl-tRNA synthetase
[EC:6.1.1.9]
Length=1042
Score = 186 bits (473), Expect = 2e-47, Method: Composition-based stats.
Identities = 91/182 (50%), Positives = 122/182 (67%), Gaps = 18/182 (9%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
+ +N +RHD GR +F++KVW+WKE++G+ I Q RR+G S+ W FTLD +S AV
Sbjct 123 QTDNVTRHDLGRDNFLKKVWEWKEKHGSAICSQFRRLGCSLDWSREFFTLDENMSKAVNN 182
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AF++LFN+G I+R+ RLVSW S L+TALSDIEVD + +P +RIPG++ VEVG LWH
Sbjct 183 AFIQLFNQGYIFRKTRLVSWCSYLRTALSDIEVDLLEVNKPSRIRIPGYEKTVEVGVLWH 242
Query 122 FVYKVEGGGE----------------IEVATTRVETMLGDVAVAVHPSDPRYQA--GPSS 163
F Y +E G+ I VATTR+ETMLGDVA+AV+P D RY++ G
Sbjct 243 FSYPLEQHGQTHEWHFMPENITNIEKITVATTRIETMLGDVAIAVNPKDNRYKSLIGKYC 302
Query 164 LH 165
LH
Sbjct 303 LH 304
> hsa:7407 VARS, G7A, VARS1, VARS2; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1264
Score = 184 bits (468), Expect = 9e-47, Method: Compositional matrix adjust.
Identities = 92/173 (53%), Positives = 121/173 (69%), Gaps = 5/173 (2%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
+E+ SRH GR+ F+++VW+WKE+ G+RI QL+++GSS+ WD CFT+D KLSAAV E
Sbjct 398 REQGLSRHQLGREAFLQEVWKWKEEKGDRIYHQLKKLGSSLDWDRACFTMDPKLSAAVTE 457
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AFVRL G+IYR RLV+WS L +A+SDIEVD + + + +PG+ VE G L
Sbjct 458 AFVRLHEEGIIYRSTRLVNWSCTLNSAISDIEVDKKELTGRTLLSVPGYKEKVEFGVLVS 517
Query 122 FVYKVEGGG---EIEVATTRVETMLGDVAVAVHPSDPRYQ--AGPSSLHFYFS 169
F YKV+G E+ VATTR+ETMLGDVAVAVHP D RYQ G + +H + S
Sbjct 518 FAYKVQGSDSDEEVVVATTRIETMLGDVAVAVHPKDTRYQHLKGKNVIHPFLS 570
> mmu:22321 Vars, Bat6, D17H6S56E, G7a, Vars2; valyl-tRNA synthetase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1263
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 92/173 (53%), Positives = 118/173 (68%), Gaps = 5/173 (2%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
KE +RH GR+ F+E+VW+WK + G+RI QL+++GSS+ WD CFT+D KLSA V E
Sbjct 397 KERGLNRHQLGREAFLEEVWKWKAEKGDRIYHQLKKLGSSLDWDRACFTMDPKLSATVTE 456
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AFVRL G+IYR RLV+WS L +A+SDIEVD + + + +PG+ VE G L
Sbjct 457 AFVRLHEEGVIYRSTRLVNWSCTLNSAISDIEVDKKELTGRTLLPVPGYKEKVEFGVLVS 516
Query 122 FVYKVEGGG---EIEVATTRVETMLGDVAVAVHPSDPRYQ--AGPSSLHFYFS 169
F YKV+G E+ VATTR+ETMLGDVAVAVHP DPRYQ G +H + S
Sbjct 517 FAYKVQGSDSDEEVVVATTRIETMLGDVAVAVHPKDPRYQHLKGKCVVHPFLS 569
> xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1243
Score = 182 bits (461), Expect = 6e-46, Method: Compositional matrix adjust.
Identities = 90/158 (56%), Positives = 114/158 (72%), Gaps = 1/158 (0%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
+E K+RHD GR++F+E+VW+WK + G+RI QLR +GSS+ WD CFT+D KLS AV E
Sbjct 379 RERGKNRHDLGRENFIEEVWKWKREKGDRIYHQLRILGSSLDWDRACFTMDPKLSFAVQE 438
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AF+RL G+IYR +RLV+WS L +A+SDIEVD + + + +PG+ VE G L
Sbjct 439 AFIRLHEAGIIYRSKRLVNWSCTLNSAISDIEVDKKELSGRTLLPVPGYKQGVEFGVLVS 498
Query 122 FVYKV-EGGGEIEVATTRVETMLGDVAVAVHPSDPRYQ 158
F YKV E G EI VATTRVETMLGD AVAVHP D RY+
Sbjct 499 FAYKVQETGEEIVVATTRVETMLGDTAVAVHPQDQRYK 536
> tpv:TP04_0740 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1010
Score = 179 bits (455), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 90/160 (56%), Positives = 112/160 (70%), Gaps = 3/160 (1%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
K+EN RHD GR FVEKV++W ++YG+ I +QL+R+G+S+ W FT+D S AV+E
Sbjct 123 KDENLKRHDLGRTKFVEKVFEWNDKYGSNIKNQLKRLGASLDWTREVFTMDQPRSTAVIE 182
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AFVRL++ G IYR RLVSW L TALSDIEV+ I P + IPG+D VEVG LW
Sbjct 183 AFVRLYDSGHIYRNTRLVSWCPYLSTALSDIEVEPMEITSPTFITIPGYDSSVEVGSLWV 242
Query 122 FVYKVEGGGE---IEVATTRVETMLGDVAVAVHPSDPRYQ 158
F Y V G E + VATTR+ETMLGDVAVAV+P D RY+
Sbjct 243 FQYPVMVGSETRYLPVATTRLETMLGDVAVAVNPDDARYK 282
> dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1057
Score = 176 bits (447), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 82/159 (51%), Positives = 115/159 (72%), Gaps = 1/159 (0%)
Query 1 LKEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVV 60
L+E+ KSR DF R++F+++VW WK + G+ I QLR++G+S+ W CFT+D S+AV
Sbjct 186 LREQGKSRQDFSREEFLKQVWMWKNEKGDEIYHQLRKLGASLDWSRACFTMDPAFSSAVT 245
Query 61 EAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLW 120
EAFVRL + GLIYR E L++WS L++A+SDIEVD++ + + +PG+ VE G +
Sbjct 246 EAFVRLCDSGLIYRAEGLINWSCTLESAISDIEVDSKELPGRTLLSVPGYQQKVEFGTMV 305
Query 121 HFVYKVEGG-GEIEVATTRVETMLGDVAVAVHPSDPRYQ 158
F Y +EG GE+ V+TTR ETMLGDVA+AVHP DPRY+
Sbjct 306 TFAYPLEGQEGEVAVSTTRPETMLGDVAIAVHPDDPRYK 344
> cel:Y87G2A.5 vrs-2; Valyl tRNA Synthetase family member (vrs-2);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1050
Score = 168 bits (426), Expect = 8e-42, Method: Compositional matrix adjust.
Identities = 80/172 (46%), Positives = 111/172 (64%), Gaps = 3/172 (1%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
+E +RHD GR F ++VW WK + G+ I DQ R++G+SV WD FT+D K+ AV E
Sbjct 181 RERGLTRHDLGRDRFNQEVWHWKNEKGDVIYDQFRKLGASVDWDRAVFTMDPKMCRAVTE 240
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AF+R+ G IYR RLV+WS L++A+SDIEVD + + + +PG+D +E G L
Sbjct 241 AFIRMHESGTIYRSNRLVNWSCALRSAISDIEVDKKELTGSTLIAVPGYDKKIEFGVLNS 300
Query 122 FVYKVEGGG-EIEVATTRVETMLGDVAVAVHPSDPRYQ--AGPSSLHFYFSS 170
F YK++G EI V+TTR+ETMLGD VAVHP D RY+ G +H + +
Sbjct 301 FAYKIQGSDEEIVVSTTRIETMLGDSGVAVHPDDQRYKHLVGKQCIHPFIPT 352
> mmu:68915 Vars2, 1190004I24Rik, Vars2l, mKIAA1885; valyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.9); K01873
valyl-tRNA synthetase [EC:6.1.1.9]
Length=1060
Score = 164 bits (414), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 79/158 (50%), Positives = 106/158 (67%), Gaps = 2/158 (1%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
KE+ RH+ R+DF+ VWQWK + G I +QL +G+S+ WD CFT+D+ SAAV E
Sbjct 200 KEQRVRRHELSREDFLRAVWQWKHEKGGEIYEQLCALGASLDWDRECFTMDAGSSAAVTE 259
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AFVRL+N GL+YR +LV+WS L++A+SDIEV++ + +++PG PV G L
Sbjct 260 AFVRLYNSGLLYRNRQLVNWSCTLRSAISDIEVESRPLPGRTVLQLPGCPTPVSFGLLAS 319
Query 122 FVYKVEG--GGEIEVATTRVETMLGDVAVAVHPSDPRY 157
+ V+G EI V TTR ET+ GDVAVAVHP DPRY
Sbjct 320 VAFPVDGEPDTEIVVGTTRPETLPGDVAVAVHPDDPRY 357
> hsa:57176 VARS2, MGC138259, MGC142165, VARS2L, VARSL; valyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.9); K01873
valyl-tRNA synthetase [EC:6.1.1.9]
Length=923
Score = 159 bits (402), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 77/158 (48%), Positives = 102/158 (64%), Gaps = 2/158 (1%)
Query 2 KEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVE 61
KE RH+ R+ F+ +VWQWKE G I +QLR +G+S+ WD CFT+D S AV E
Sbjct 60 KERGVRRHELSREAFLREVWQWKEAKGGEICEQLRALGASLDWDRECFTMDVGSSVAVTE 119
Query 62 AFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWH 121
AFVRL+ GL+YR +LV+WS L++A+SDIEV+ + +R+PG PV G L+
Sbjct 120 AFVRLYKAGLLYRNHQLVNWSCALRSAISDIEVENRPLPGHTQLRLPGCPTPVSFGLLFS 179
Query 122 FVYKVEG--GGEIEVATTRVETMLGDVAVAVHPSDPRY 157
+ V+G E+ V TTR ET+ GDVAVAVHP D RY
Sbjct 180 VAFPVDGEPDAEVVVGTTRPETLPGDVAVAVHPDDSRY 217
> sce:YGR094W VAS1; Mitochondrial and cytoplasmic valyl-tRNA synthetase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1104
Score = 158 bits (399), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 76/157 (48%), Positives = 110/157 (70%), Gaps = 1/157 (0%)
Query 3 EENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEA 62
++ K+RHD+GR+ FV KVW+WKE+Y +RI +Q++++G+S W FTL +L+ +V EA
Sbjct 245 KDRKTRHDYGREAFVGKVWEWKEEYHSRIKNQIQKLGASYDWSREAFTLSPELTKSVEEA 304
Query 63 FVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWHF 122
FVRL + G+IYR RLV+WS L TA+S++EV+ + ++ + +PG+D VE G L F
Sbjct 305 FVRLHDEGVIYRASRLVNWSVKLNTAISNLEVENKDVKSRTLLSVPGYDEKVEFGVLTSF 364
Query 123 VYKVEGGGE-IEVATTRVETMLGDVAVAVHPSDPRYQ 158
Y V G E + +ATTR ET+ GD AVAVHP D RY+
Sbjct 365 AYPVIGSDEKLIIATTRPETIFGDTAVAVHPDDDRYK 401
> dre:565521 rCG46902-like
Length=2172
Score = 146 bits (368), Expect = 4e-35, Method: Composition-based stats.
Identities = 62/133 (46%), Positives = 94/133 (70%), Gaps = 0/133 (0%)
Query 1 LKEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVV 60
++E +RHD GR++F+++VW+WK + G+RI QL+++GSS+ WD CFT+D KLS AV
Sbjct 403 MRERKMTRHDLGRENFIKEVWKWKNEKGDRIYHQLKKLGSSLDWDRACFTMDDKLSFAVQ 462
Query 61 EAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLW 120
EAF+R+ G+IYR +RLV+WS L +A+SDIEVD + + + +PG+ VE G L
Sbjct 463 EAFIRMHEEGVIYRSKRLVNWSCTLNSAISDIEVDKKELTGRTLLPVPGYKDKVEFGVLV 522
Query 121 HFVYKVEGGGEIE 133
F YK+EG +++
Sbjct 523 SFSYKIEGSDDLD 535
> ath:AT5G16715 EMB2247 (embryo defective 2247); ATP binding /
aminoacyl-tRNA ligase/ nucleotide binding / valine-tRNA ligase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=970
Score = 145 bits (367), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 76/158 (48%), Positives = 104/158 (65%), Gaps = 17/158 (10%)
Query 1 LKEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVV 60
L E R D GR +F ++VW+WKE+YG IT+Q++R+G+S W FTLD +LS AVV
Sbjct 161 LASEGIKRVDLGRDEFTKRVWEWKEKYGGTITNQIKRLGASCDWSRERFTLDEQLSRAVV 220
Query 61 EAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLW 120
EAFV+L ++GLIY+ +V+WS L+TA+SD+EV E E+P GFL+
Sbjct 221 EAFVKLHDKGLIYQGSYMVNWSPNLQTAVSDLEV--EYSEEP--------------GFLY 264
Query 121 HFVYKVEGGGE-IEVATTRVETMLGDVAVAVHPSDPRY 157
H Y+V G + + +ATTR ET+ GDVA+AVHP D RY
Sbjct 265 HIKYRVAGSPDFLTIATTRPETLFGDVALAVHPEDDRY 302
> eco:b4258 valS, ECK4251, JW4215; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=951
Score = 145 bits (366), Expect = 7e-35, Method: Composition-based stats.
Identities = 73/163 (44%), Positives = 101/163 (61%), Gaps = 23/163 (14%)
Query 3 EENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEA 62
EE K+RHD+GR+ F++K+W+WK + G IT Q+RR+G+SV W+ FT+D LS AV E
Sbjct 97 EEGKTRHDYGREAFIDKIWEWKAESGGTITRQMRRLGNSVDWERERFTMDEGLSNAVKEV 156
Query 63 FVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRIPGFDFPVEVGFLWHF 122
FVRL+ LIYR +RLV+W L+TA+SD+EV+ + G +WH
Sbjct 157 FVRLYKEDLIYRGKRLVNWDPKLRTAISDLEVENRESK----------------GSMWHI 200
Query 123 VYKVEGGGE-------IEVATTRVETMLGDVAVAVHPSDPRYQ 158
Y + G + + VATTR ET+LGD VAV+P DPRY+
Sbjct 201 RYPLADGAKTADGKDYLVVATTRPETLLGDTGVAVNPEDPRYK 243
> cel:ZC513.4 vrs-1; Valyl tRNA Synthetase family member (vrs-1);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=918
Score = 95.5 bits (236), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 63/162 (38%), Positives = 86/162 (53%), Gaps = 5/162 (3%)
Query 1 LKEENKSRHDFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVV 60
L + K R + R++FV++ W E+ + I QL R+G+S+ W +TLDSK S AV
Sbjct 58 LAKNGKRRLEMTREEFVKECHLWGEKCSSEIRQQLTRMGASLDWTQSYYTLDSKFSEAVT 117
Query 61 EAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRI---PGFDFPVEVG 117
+AF L LI R +RLV W L + LS EV+ + + + G V+ G
Sbjct 118 KAFCILEQEDLITRGKRLVHWCPTLSSTLSSQEVNRIDVPSDGYINLSSSSGTRRRVKFG 177
Query 118 FLWHFVYK-VEGGGE-IEVATTRVETMLGDVAVAVHPSDPRY 157
+ Y V+ E IEV TTR ET+ DVAVAVHP D RY
Sbjct 178 QMHVIRYNLVDSESEFIEVGTTRPETLFADVAVAVHPEDDRY 219
> dre:100001219 iars2, si:ch211-214p16.3, si:zc214p16.3; isoleucine-tRNA
synthetase 2, mitochondrial (EC:6.1.1.5); K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=983
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 42/161 (26%), Positives = 73/161 (45%), Gaps = 15/161 (9%)
Query 1 LKEENKSRHDFGRKDFV-----EKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKL 55
L E K+ D G + +K ++ E+ +R +R G WD+ +T D K
Sbjct 129 LPIELKALGDLGTNELTPVQIRQKAREFAEKAISRQRAAFQRWGVMADWDNCYYTFDGKY 188
Query 56 SAAVVEAFVRLFNRGLIYREERLVSWSSCLKTAL--SDIEVDTETIEQPQTVRIPGFDFP 113
AA ++ F + N+GLIY++ + V WS +TAL +++E + E + + + P P
Sbjct 189 EAAQLKVFQEMHNKGLIYQDYKPVFWSPSSRTALAEAELEYNPEHVSRSLYITFPLLTLP 248
Query 114 VEVGFLWHFVYKVEGGGEIE--VATTRVETMLGDVAVAVHP 152
+ K EG + + TT+ T+ + AV P
Sbjct 249 AK------LTAKAEGLSNVSALIWTTQPWTIPANQAVCYMP 283
> tgo:TGME49_016500 tRNA synthetase, putative (EC:6.1.1.9 6.1.1.4)
Length=1762
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 31 ITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALS 90
I Q RR+G S W +TLD V AFV+LF + I + L W +TAL+
Sbjct 303 IERQQRRLGCSCDWSRRVYTLDGPYCDLVTRAFVQLFRQKYIRKGTYLTLWDCGAQTALA 362
Query 91 DIEV 94
D EV
Sbjct 363 DFEV 366
> mmu:381314 Iars2, 2010002H18Rik, C79125, MGC63429; isoleucine-tRNA
synthetase 2, mitochondrial (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1012
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query 37 RVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTAL--SDIEV 94
R G W++ +T D K A + F +++ +GL+YR + V WS +TAL +++E
Sbjct 201 RWGVMADWNNCYYTFDPKYEAKQLRVFYQMYEKGLVYRSYKPVYWSPSSRTALAEAELEY 260
Query 95 DTETIEQPQTVRIPGFDFPVEV 116
+ E + + VR P P ++
Sbjct 261 NPEHVSRSIYVRFPLLRPPPKL 282
> ath:AT5G49030 OVA2; OVA2 (ovule abortion 2); ATP binding / aminoacyl-tRNA
ligase/ catalytic/ isoleucine-tRNA ligase/ nucleotide
binding; K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=955
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 33 DQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDI 92
+ +R G W++ TLD + AA +E F ++ +G IYR + V WS +TAL++
Sbjct 237 ESFKRFGVWADWNNPYLTLDPEYEAAQIEVFGQMALKGYIYRGRKPVHWSPSSRTALAEA 296
Query 93 EVD 95
E++
Sbjct 297 ELE 299
> hsa:55699 IARS2, FLJ10326, ILERS; isoleucyl-tRNA synthetase
2, mitochondrial (EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase
[EC:6.1.1.5]
Length=1012
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 58/120 (48%), Gaps = 10/120 (8%)
Query 37 RVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTAL--SDIEV 94
R G W++ +T D K A + F +++++GL+YR + V WS +TAL +++E
Sbjct 201 RWGIMADWNNCYYTFDGKYEAKQLRTFYQMYDKGLVYRSYKPVFWSPSSRTALAEAELEY 260
Query 95 DTETIEQPQTVRIPGFDFPVEVGFLWHFVYKVEGGGEIEVA--TTRVETMLGDVAVAVHP 152
+ E + + V+ P ++ L ++G + + TT+ T+ + AV P
Sbjct 261 NPEHVSRSIYVKFPLLKPSPKLASL------IDGSSPVSILVWTTQPWTIPANEAVCYMP 314
> cpv:cgd3_3840 isoleucine-tRNA synthetase ; K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1100
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 35/131 (26%), Positives = 60/131 (45%), Gaps = 20/131 (15%)
Query 26 QYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCL 85
+Y + RVG + +D+ T+D V F +LF++ L+YR R++ +S+
Sbjct 128 RYTQEWKSTVERVGRLIDFDNGYKTMDLNFMQTVWWVFKQLFDKNLVYRAYRVMPYSTAC 187
Query 86 KTALSDIE--VDTETIEQPQT-VRIPGFDFPVEVGFL-WHFVYKVEGGGEIEVATTRVET 141
T LS+ E ++ + + P + P + P + FL W TT T
Sbjct 188 ATPLSNFECNLNYKDVNDPSVIITFPLLEDP-NIQFLAW---------------TTTPWT 231
Query 142 MLGDVAVAVHP 152
+ +VA+AVHP
Sbjct 232 LPSNVAIAVHP 242
> tgo:TGME49_007640 isoleucine-tRNA synthetase, putative (EC:6.1.1.5);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1103
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 49/92 (53%), Gaps = 3/92 (3%)
Query 7 SRHD---FGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAF 63
SRH+ G + +K +Y + + R+G + +D+ TLD++ +V F
Sbjct 111 SRHEIVEMGIDKYNDKCRSIVMRYASEWRSVIERMGRWIDFDNDYKTLDTRFMESVWWVF 170
Query 64 VRLFNRGLIYREERLVSWSSCLKTALSDIEVD 95
+LF++GL+YR +++ +S+ T LS+ E +
Sbjct 171 KQLFDKGLVYRANKIMPFSTACSTPLSNFEAN 202
> xla:443989 lars2, MGC80460; leucyl-tRNA synthetase 2, mitochondrial
(EC:6.1.1.4); K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=900
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 37/73 (50%), Gaps = 0/73 (0%)
Query 23 WKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWS 82
W E+ + +QL R+G +WD T F++++ GL Y++E +V+W
Sbjct 145 WTERNIKHMREQLERIGLCFNWDREITTCSEDYYKWTQYLFLKMYEAGLAYQKEAMVNWD 204
Query 83 SCLKTALSDIEVD 95
+T L++ +VD
Sbjct 205 PVDQTVLANEQVD 217
> hsa:23395 LARS2, KIAA0028, LEURS, MGC26121; leucyl-tRNA synthetase
2, mitochondrial (EC:6.1.1.4); K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=903
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 0/73 (0%)
Query 23 WKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWS 82
W + + QL R+G SWD T F++L+ GL Y++E LV+W
Sbjct 149 WTQSNIKHMRKQLDRLGLCFSWDREITTCLPDYYKWTQYLFIKLYEAGLAYQKEALVNWD 208
Query 83 SCLKTALSDIEVD 95
+T L++ +VD
Sbjct 209 PVDQTVLANEQVD 221
> tpv:TP02_0541 hypothetical protein
Length=1221
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 32/55 (58%), Gaps = 0/55 (0%)
Query 44 WDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTET 98
WD++ T S+ V+ F++ F+ GLIY+ E +SS K+ L+D EV +T
Sbjct 300 WDYYYATYHSRFEKQVMTTFMKFFDSGLIYKAELPQIYSSKSKSVLADSEVIQKT 354
> mmu:102436 Lars2, AI035546, Kiaa0028, LEURS; leucyl-tRNA synthetase,
mitochondrial (EC:6.1.1.4); K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=902
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 0/73 (0%)
Query 23 WKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWS 82
W + + QL R+G SWD T F++L+ GL Y++E LV+W
Sbjct 148 WTQSNIKHMRKQLDRLGLCFSWDREITTCLPDYYKWTQYLFIKLYEAGLAYQKEALVNWD 207
Query 83 SCLKTALSDIEVD 95
+T L++ +V+
Sbjct 208 PVDQTVLANEQVN 220
> sce:YLR382C NAM2, MSL1; LeuRS; mitochondrial leucyl-tRNA synthetase
(EC:6.1.1.4); K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=894
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 36/75 (48%), Gaps = 0/75 (0%)
Query 23 WKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWS 82
W ++ Q++ + ++ WD T D + F++LF GL YR+E ++W
Sbjct 114 WTRDNIAKMKQQMQSMLANFDWDREITTCDPEYYKFTQWIFLKLFENGLAYRKEAEINWD 173
Query 83 SCLKTALSDIEVDTE 97
T L++ +VD +
Sbjct 174 PVDMTVLANEQVDAQ 188
> ath:AT4G04350 EMB2369 (EMBRYO DEFECTIVE 2369); ATP binding /
aminoacyl-tRNA ligase/ leucine-tRNA ligase/ nucleotide binding
(EC:6.1.1.4); K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=973
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 33/66 (50%), Gaps = 0/66 (0%)
Query 29 NRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTA 88
+R QL+ +G S WD T + F++L+ +GL Y+ E V+W L T
Sbjct 189 DRFRLQLKSLGFSYDWDRELSTTEPDYYKWTQWIFLQLYKKGLAYQAEVPVNWCPALGTV 248
Query 89 LSDIEV 94
L++ EV
Sbjct 249 LANEEV 254
> dre:100126021 zgc:165522 (EC:6.1.1.4); K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=891
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 33/73 (45%), Gaps = 0/73 (0%)
Query 23 WKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWS 82
W + QL +G +WD T F+++F GL Y++E LV+W
Sbjct 136 WTRSNIESMRTQLDSLGLCFNWDREITTCLPDYYKWTQYLFIKMFEAGLAYQKEALVNWD 195
Query 83 SCLKTALSDIEVD 95
+T L+D +VD
Sbjct 196 PVDQTVLADEQVD 208
> tgo:TGME49_077030 isoleucyl-tRNA synthetase, putative (EC:6.1.1.4
6.1.1.5); K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1988
Score = 37.7 bits (86), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 0/60 (0%)
Query 36 RRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVD 95
+R G W+ T D A +E F R++ G +Y R V WS +TAL+D EV+
Sbjct 622 QRFGVWAHWEKPYLTYDRNYEATELEMFRRMWRLGFVYEGVRPVFWSPSARTALADAEVE 681
> tgo:TGME49_066730 leucyl-tRNA synthetase, putative (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1465
Score = 37.0 bits (84), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 32/66 (48%), Gaps = 0/66 (0%)
Query 29 NRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTA 88
+R QLRR+G V W T + FV++ RGL+Y + V+W L T
Sbjct 384 DRFRQQLRRLGVCVDWRREVNTSSPEFYRWTQWIFVQMMKRGLVYEKLANVNWCEGLGTV 443
Query 89 LSDIEV 94
L++ EV
Sbjct 444 LANEEV 449
> eco:b0026 ileS, ECK0027, ilvS, JW0024; isoleucyl-tRNA synthetase
(EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=938
Score = 37.0 bits (84), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 37 RVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVD 95
R+G W H T+D K A ++ A ++ G +++ + V W ++AL++ EV+
Sbjct 143 RLGVLGDWSHPYLTMDFKTEANIIRALGKIIGNGHLHKGAKPVHWCVDCRSALAEAEVE 201
> cel:C25A1.7 irs-2; Isoleucyl tRNA Synthetase family member (irs-2);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=970
Score = 35.4 bits (80), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 0/63 (0%)
Query 33 DQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDI 92
+ RR G + W+ T AA ++ F +L + L+YR + V WS TAL++
Sbjct 176 NAFRRWGVTADWEKPYVTKSPSYVAAQLDIFAKLVEQKLVYRSFKPVYWSPSSNTALAES 235
Query 93 EVD 95
E++
Sbjct 236 ELE 238
> bbo:BBOV_IV002370 21.m02840; leucyl-tRNA synthetase (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=782
Score = 35.4 bits (80), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 29/65 (44%), Gaps = 0/65 (0%)
Query 30 RITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTAL 89
R QL+R+G + W T D F + G+ YR +LV+W L T L
Sbjct 185 RFKHQLKRMGFAFDWSRELATSDPGYYKWTQWMFQEFYRSGIAYRANQLVNWCPDLGTVL 244
Query 90 SDIEV 94
++ EV
Sbjct 245 ANEEV 249
> sce:YPL040C ISM1; Ism1p (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1002
Score = 33.5 bits (75), Expect = 0.35, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 44 WDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVD 95
W+ T+D + F ++ RGLI R+ + V W + +TAL++ E++
Sbjct 187 WETPYLTMDKDYEINQLNIFKEMYERGLIKRQNKPVYWGTETRTALAEGELE 238
> hsa:3376 IARS, FLJ20736, IARS1, ILERS, ILRS, IRS, PRO0785; isoleucyl-tRNA
synthetase (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1262
Score = 33.1 bits (74), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 37 RVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIE 93
R+G + +D+ TL + +V F +L+++GL+YR +++ +S+ T LS+ E
Sbjct 137 RLGRWIDFDNDYKTLYPQFMESVWWVFKQLYDKGLVYRGVKVMPFSTACNTPLSNFE 193
> hsa:5314 PKHD1, ARPKD, DKFZp686C01112, FCYT, FLJ46150, TIGM1;
polycystic kidney and hepatic disease 1 (autosomal recessive)
Length=4074
Score = 32.7 bits (73), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 48 CFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIEVDTETIEQPQTVRI 107
C + + ++ +F LF + R+E L+ SSC T ++ E++ ET QP TV+I
Sbjct 1846 CLFVPDHWAESMFPSFSGLFISPKLERDEVLIYNSSCNITMETEAEMECETPNQPITVKI 1905
> mmu:105148 Iars, 2510016L12Rik, AI327140, AU044614, E430001P04Rik,
ILRS; isoleucine-tRNA synthetase (EC:6.1.1.5); K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1262
Score = 32.3 bits (72), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 37 RVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIE 93
R+G + +D+ TL + +V F +L+++GL+YR +++ +S+ T LS+ E
Sbjct 137 RLGRWIDFDNDYKTLYPQFMESVWWVFKQLYDKGLVYRGVKVMPFSTACGTPLSNFE 193
> cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family
member ; K12849 pre-mRNA-splicing factor 38A
Length=261
Score = 32.3 bits (72), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 39/86 (45%), Gaps = 9/86 (10%)
Query 37 RVGSSVSWDHFCFTLDSK--LSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIEV 94
RV SS+ W CF LDS+ L AV+ ++ G Y +R + CL L I
Sbjct 21 RVFSSIYWKGECFALDSETILDKAVLLDYI-----GTTYGGDRKATPFLCLLVKLLQIRP 75
Query 95 DTETIEQPQTVRIPGFDFPVEVGFLW 120
TE + + + P F + +G ++
Sbjct 76 STEIV--LEYINNPRFKYLTALGIVY 99
> pfa:PFC0470w valine-tRNA ligase, putative (EC:6.1.1.9); K01873
valyl-tRNA synthetase [EC:6.1.1.9]
Length=1367
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 17/91 (18%), Positives = 42/91 (46%), Gaps = 0/91 (0%)
Query 10 DFGRKDFVEKVWQWKEQYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNR 69
+ GR+ ++ ++ +W+E + L+ + + + T+D + + +AF L+
Sbjct 325 ELGREKYINEIKKWQENLKENMLKDLQNMNITYNEKMLFNTMDKGMIDLINKAFYILYKN 384
Query 70 GLIYREERLVSWSSCLKTALSDIEVDTETIE 100
+I V + LKTA+ +++ + E
Sbjct 385 NMIINRLYPVYYCKELKTAIPKMDIQFKNAE 415
> pfa:PF13_0179 isoleucine-tRNA ligase, putative (EC:6.1.1.5);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1272
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 20/92 (21%), Positives = 40/92 (43%), Gaps = 4/92 (4%)
Query 26 QYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCL 85
+Y ++R+G + + + T+D +V F L+ IY+ +++ +S
Sbjct 209 KYSKEWVATVQRIGRWIDFKNDYKTMDKSFMESVWWVFSELYKNNYIYKSFKVMPYSCKC 268
Query 86 KTALSDIEVDTETIEQPQTVRIPGF----DFP 113
T +S+ E++ + P I F DFP
Sbjct 269 NTPISNFELNLNYKDTPDPSIIISFILCSDFP 300
> dre:334393 iars, fi46h05, wu:fi46h05, zgc:63790; isoleucyl-tRNA
synthetase (EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase
[EC:6.1.1.5]
Length=1271
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/91 (23%), Positives = 46/91 (50%), Gaps = 6/91 (6%)
Query 26 QYGNRITDQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCL 85
+Y + + +RR+G + + + TL V F +L++ GL+YR +++ +S+
Sbjct 126 RYADEWENSVRRMGRWIDFRNDYKTLYPWFMETVWWVFKQLYDSGLVYRGVKVMPFSTAC 185
Query 86 KTALSDIEVDT--ETIEQPQTVRIPGFDFPV 114
T LS+ E + + ++ P + +FP+
Sbjct 186 NTPLSNFEANQNYKDVQDPSVI----VNFPL 212
> pfa:PF08_0011 leucine-tRNA ligase; K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=1481
Score = 30.8 bits (68), Expect = 2.5, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 32/65 (49%), Gaps = 0/65 (0%)
Query 33 DQLRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDI 92
+QL ++G +W+ T D ++LF L Y++ V+WS+ L+ +S+
Sbjct 213 NQLIKLGFLFNWESEINTCDENYYKWTQWIIIQLFLNNLSYKKRSYVNWSNELRCVISND 272
Query 93 EVDTE 97
E+ E
Sbjct 273 ELRNE 277
> xla:403361 iars, MGC68929; isoleucyl-tRNA synthetase; K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1259
Score = 30.0 bits (66), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 44/93 (47%), Gaps = 18/93 (19%)
Query 63 FVRLFNRGLIYREERLVSWSSCLKTALSDIEV--DTETIEQPQT-VRIPGFDFPVEVGFL 119
F +L+++GL+YR +++ +S+ T LS+ E + + ++ P V P D P
Sbjct 163 FKQLYDKGLVYRGVKVMPFSTACNTPLSNFESHQNYKDVQDPSVIVTFPLLDDP------ 216
Query 120 WHFVYKVEGGGEIEVATTRVETMLGDVAVAVHP 152
G + TT T+ ++A+ V+P
Sbjct 217 ---------GISLVAWTTTPWTLPSNLALCVNP 240
> dre:556241 very large inducible GTPase-1-like
Length=1679
Score = 30.0 bits (66), Expect = 3.7, Method: Composition-based stats.
Identities = 26/93 (27%), Positives = 52/93 (55%), Gaps = 11/93 (11%)
Query 9 HDFGRKDFVEKVWQWK-EQYGNRITDQL---RRVGSSVSWDHFCFTLDSKLSAAVVEAFV 64
+DF +K+ E + Q K + N ITD L + + ++V +++F F+ + L V +A++
Sbjct 931 YDF-KKNMAEIIAQCKCNTHVNNITDFLEWTKSLWNAVKYENFIFSFRNSL---VADAYM 986
Query 65 RL---FNRGLIYREERLVSWSSCLKTALSDIEV 94
RL +NR ++ + SW + +T +S+ E+
Sbjct 987 RLCAEYNRWEWSFKKEMHSWLTQAETRVSNFEI 1019
> ath:AT4G10320 isoleucyl-tRNA synthetase, putative / isoleucine--tRNA
ligase, putative (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1190
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 34/68 (50%), Gaps = 0/68 (0%)
Query 35 LRRVGSSVSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREERLVSWSSCLKTALSDIEV 94
+ R G + + + T+D +V F +L+ + L+YR +++ +S+ KT LS+ E
Sbjct 136 ITRCGRWIDFKNDYKTMDLPFMESVWWVFSQLWEKNLVYRGFKVMPYSTGCKTPLSNFEA 195
Query 95 DTETIEQP 102
E P
Sbjct 196 GQNYKEVP 203
> ath:AT3G55400 OVA1; OVA1 (OVULE ABORTION 1); ATP binding / aminoacyl-tRNA
ligase/ methionine-tRNA ligase/ nucleotide binding;
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=533
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 42 VSWDHFCFTLDSKLSAAVVEAFVRLFNRGLIYREE 76
+++D F T D K A V E + R+F G IYR +
Sbjct 153 IAYDKFIRTTDPKHEAIVKEFYARVFANGDIYRAD 187
Lambda K H
0.321 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4287611064
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40