bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3597_orf1
Length=204
Score E
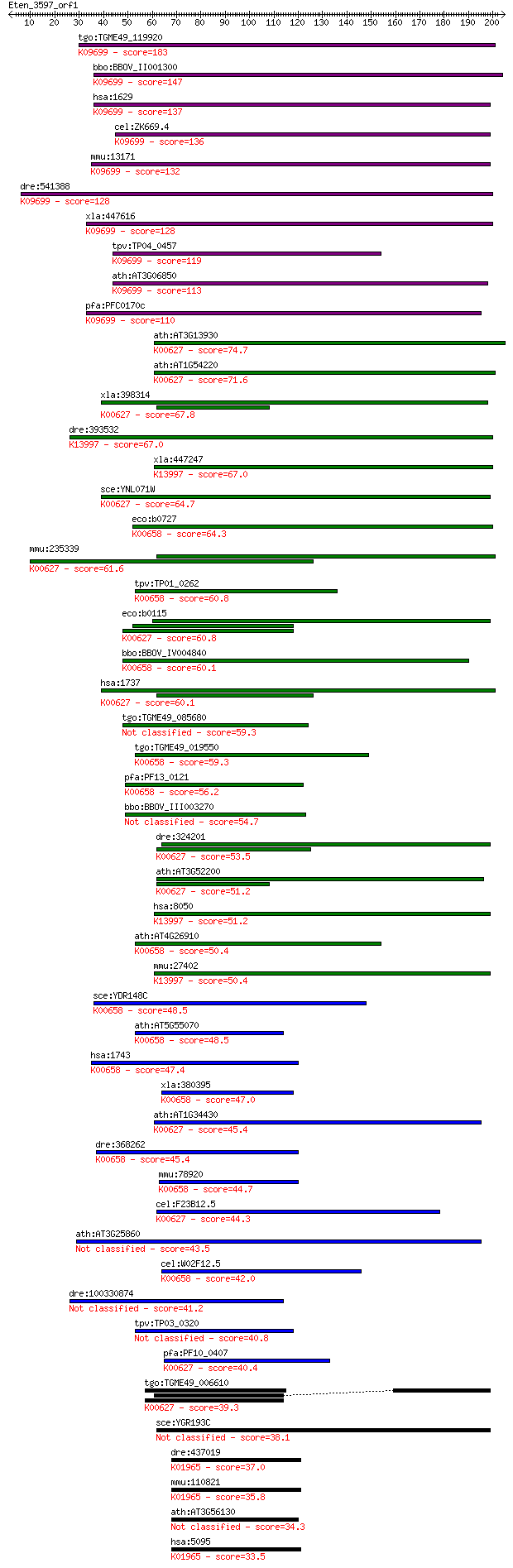
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_119920 dihydrolipoamide branched chain transacylase... 183 3e-46
bbo:BBOV_II001300 18.m06098; lipoamide acyltransferase compone... 147 2e-35
hsa:1629 DBT, BCATE2, E2, E2B, MGC9061; dihydrolipoamide branc... 137 3e-32
cel:ZK669.4 hypothetical protein; K09699 2-oxoisovalerate dehy... 136 5e-32
mmu:13171 Dbt, D3Wsu60e; dihydrolipoamide branched chain trans... 132 6e-31
dre:541388 dbt, im:7147214, zgc:103768; dihydrolipoamide branc... 128 1e-29
xla:447616 dbt, MGC85493; dihydrolipoamide branched chain tran... 128 1e-29
tpv:TP04_0457 lipoamide transferase (EC:2.3.1.-); K09699 2-oxo... 119 7e-27
ath:AT3G06850 BCE2; BCE2; acetyltransferase/ alpha-ketoacid de... 113 5e-25
pfa:PFC0170c dihydrolipoamide acyltransferase, putative (EC:2.... 110 3e-24
ath:AT3G13930 dihydrolipoamide S-acetyltransferase, putative (... 74.7 2e-13
ath:AT1G54220 dihydrolipoamide S-acetyltransferase, putative (... 71.6 2e-12
xla:398314 dlat; dihydrolipoamide S-acetyltransferase (EC:2.3.... 67.8 3e-11
dre:393532 pdhx, MGC66110, zgc:66110, zgc:85977; pyruvate dehy... 67.0 4e-11
xla:447247 pdhx, MGC86218; pyruvate dehydrogenase complex, com... 67.0 5e-11
sce:YNL071W LAT1, ODP2, PDA2; Dihydrolipoamide acetyltransfera... 64.7 2e-10
eco:b0727 sucB, ECK0715, JW0716; dihydrolipoyltranssuccinase (... 64.3 3e-10
mmu:235339 Dlat, 6332404G05Rik, DLTA, PDC-E2; dihydrolipoamide... 61.6 2e-09
tpv:TP01_0262 dihydrolipoamide succinyltransferase; K00658 2-o... 60.8 3e-09
eco:b0115 aceF, ECK0114, JW0111; pyruvate dehydrogenase, dihyd... 60.8 3e-09
bbo:BBOV_IV004840 23.m06243; dihydrolipoamide succinyltransfer... 60.1 5e-09
hsa:1737 DLAT, DLTA, PDC-E2, PDCE2; dihydrolipoamide S-acetylt... 60.1 5e-09
tgo:TGME49_085680 dihydrolipoamide acyltransferase, putative (... 59.3 8e-09
tgo:TGME49_019550 dihydrolipoamide succinyltransferase compone... 59.3 9e-09
pfa:PF13_0121 dihydrolipamide succinyltransferase component of... 56.2 7e-08
bbo:BBOV_III003270 17.m07312; biotin-requiring enzyme family p... 54.7 2e-07
dre:324201 dlat, wu:fc14f10, wu:fc21f08, wu:fc86g11, wu:fj57d0... 53.5 5e-07
ath:AT3G52200 LTA3; LTA3; ATP binding / dihydrolipoyllysine-re... 51.2 2e-06
hsa:8050 PDHX, DLDBP, E3BP, OPDX, PDX1, proX; pyruvate dehydro... 51.2 2e-06
ath:AT4G26910 2-oxoacid dehydrogenase family protein (EC:2.3.1... 50.4 4e-06
mmu:27402 Pdhx, AI481367, E3bp, Pdx1; pyruvate dehydrogenase c... 50.4 5e-06
sce:YDR148C KGD2; Dihydrolipoyl transsuccinylase, component of... 48.5 2e-05
ath:AT5G55070 2-oxoacid dehydrogenase family protein (EC:2.3.1... 48.5 2e-05
hsa:1743 DLST, DLTS; dihydrolipoamide S-succinyltransferase (E... 47.4 4e-05
xla:380395 dlst, MGC53142; dihydrolipoamide S-succinyltransfer... 47.0 4e-05
ath:AT1G34430 EMB3003 (embryo defective 3003); acyltransferase... 45.4 1e-04
dre:368262 dlst, cb60, wu:fc39d01; dihydrolipoamide S-succinyl... 45.4 1e-04
mmu:78920 Dlst, 1600017E01Rik, 4632413C10Rik, 4930529O08Rik, D... 44.7 2e-04
cel:F23B12.5 hypothetical protein; K00627 pyruvate dehydrogena... 44.3 3e-04
ath:AT3G25860 LTA2; LTA2; dihydrolipoyllysine-residue acetyltr... 43.5 4e-04
cel:W02F12.5 hypothetical protein; K00658 2-oxoglutarate dehyd... 42.0 0.001
dre:100330874 pyruvate dehydrogenase complex, component X-like 41.2 0.002
tpv:TP03_0320 hypothetical protein 40.8 0.003
pfa:PF10_0407 dihydrolipoamide acyltransferase, putative (EC:2... 40.4 0.005
tgo:TGME49_006610 biotin requiring domain-containing protein /... 39.3 0.009
sce:YGR193C PDX1; Dihydrolipoamide dehydrogenase (E3)-binding ... 38.1 0.023
dre:437019 pcca, wu:fb92g02, zgc:100925; propionyl-Coenzyme A ... 37.0 0.051
mmu:110821 Pcca, C79630; propionyl-Coenzyme A carboxylase, alp... 35.8 0.10
ath:AT3G56130 biotin/lipoyl attachment domain-containing protein 34.3 0.33
hsa:5095 PCCA; propionyl CoA carboxylase, alpha polypeptide (E... 33.5 0.55
> tgo:TGME49_119920 dihydrolipoamide branched chain transacylase,
E2 subunit, putative (EC:2.3.1.168); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=510
Score = 183 bits (465), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 102/176 (57%), Positives = 120/176 (68%), Gaps = 8/176 (4%)
Query 30 PSTRGVFTVSHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAV 89
P R + TVS P + FKLADIGEGIA VEL KW+K GD VEEM+E+CEVQSDKAAV
Sbjct 56 PGKRCLLTVSRPALAVKTFKLADIGEGIAQVELLKWHKGVGDHVEEMDELCEVQSDKAAV 115
Query 90 EITSRYSGKIVKLYAKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQPS 149
EITSR++G IVKL+ KEG V+IGAPL+DID E+ ++ P+ S+PQ
Sbjct 116 EITSRFTGTIVKLHQKEGMMVRIGAPLMDIDVEAGEDHAEEEEPETKERPAPVSEPQ--- 172
Query 150 APASSSRGAEP-----LASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYLSS 200
A AS S GAE ASPA RRFAKEKGV+L VKG+G G ITKEDVL +L S
Sbjct 173 AAASPSVGAEASSTTFSASPATRRFAKEKGVDLARVKGSGRNGLITKEDVLKFLES 228
> bbo:BBOV_II001300 18.m06098; lipoamide acyltransferase component
of branched-chain alpha-keto acid dehydrogenase complex
(EC:2.3.1.12); K09699 2-oxoisovalerate dehydrogenase E2 component
(dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=417
Score = 147 bits (372), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 79/172 (45%), Positives = 108/172 (62%), Gaps = 12/172 (6%)
Query 36 FTVSHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRY 95
F S R+ + F L+DIGEGI+ VEL +W K GD VEEME VC VQSDKAAV+ITSRY
Sbjct 22 FHRSVHRNKLTTFHLSDIGEGISEVELVRWNKNVGDEVEEMETVCTVQSDKAAVDITSRY 81
Query 96 SGKIVKLYAKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQPSAPASS- 154
+G + KLY ++G +KIG+PL+DID+ D +PA P+E +K PS P +
Sbjct 82 TGLVKKLYVEQGKLIKIGSPLMDIDAED--------DTPAVSEPTETTKSSIPSKPVAQS 133
Query 155 ---SRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYLSSGAS 203
S G A+P+VR+ AK+ GV++ V +G+ IT+EDV + +S S
Sbjct 134 FKRSHGDSVRAAPSVRQLAKQLGVDITKVVPSGSNSQITREDVEKFAASSQS 185
> hsa:1629 DBT, BCATE2, E2, E2B, MGC9061; dihydrolipoamide branched
chain transacylase E2 (EC:2.3.1.168); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=482
Score = 137 bits (344), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 75/173 (43%), Positives = 98/173 (56%), Gaps = 18/173 (10%)
Query 36 FTVSHPRHG----------IVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSD 85
F SHP H +V FKL+DIGEGI V + +WY K GDTV + + +CEVQSD
Sbjct 45 FKYSHPHHFLKTTAALRGQVVQFKLSDIGEGIREVTVKEWYVKEGDTVSQFDSICEVQSD 104
Query 86 KAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKP 145
KA+V ITSRY G I KLY D +G PL+DI++ +++ S++ P S
Sbjct 105 KASVTITSRYDGVIKKLYYNLDDIAYVGKPLVDIETEALKD--SEEDVVETPAVSHDEHT 162
Query 146 QQPSAPASSSRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYL 198
Q +G + LA+PAVRR A E + L V G+G G I KED+LNYL
Sbjct 163 HQ------EIKGRKTLATPAVRRLAMENNIKLSEVVGSGKDGRILKEDILNYL 209
> cel:ZK669.4 hypothetical protein; K09699 2-oxoisovalerate dehydrogenase
E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=448
Score = 136 bits (342), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 73/155 (47%), Positives = 100/155 (64%), Gaps = 2/155 (1%)
Query 45 IVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYA 104
+V FKL+DIGEGIA V++ +WY K GDT+ + ++VCEVQSDKAAV I+ RY G + KLY
Sbjct 30 VVQFKLSDIGEGIAEVQVKEWYVKEGDTISQFDKVCEVQSDKAAVTISCRYDGIVKKLYH 89
Query 105 KEGDTVKIGAPLIDID-SPDVEETQSQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLAS 163
+ ++G LID++ +VEE + + A+ P EA K P AP S+ + LA+
Sbjct 90 EVDGMARVGQALIDVEIEGNVEEPEQPKKEAASSSP-EAPKSSAPKAPESAHSEGKVLAT 148
Query 164 PAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYL 198
PAVRR A E + L V+GTG G + KEDVL +L
Sbjct 149 PAVRRIAIENKIKLAEVRGTGKDGRVLKEDVLKFL 183
> mmu:13171 Dbt, D3Wsu60e; dihydrolipoamide branched chain transacylase
E2 (EC:2.3.1.168); K09699 2-oxoisovalerate dehydrogenase
E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=482
Score = 132 bits (333), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 73/174 (41%), Positives = 99/174 (56%), Gaps = 18/174 (10%)
Query 35 VFTVSHPRHG----------IVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQS 84
+F S PRH +V FKL+DIGEGI V + +WY K GDTV + + +CEVQS
Sbjct 44 LFKYSQPRHSLRTAAVLQGQVVQFKLSDIGEGIREVTIKEWYVKEGDTVSQFDSICEVQS 103
Query 85 DKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASK 144
DKA+V ITSRY G I +LY D +G PLIDI++ +++ S++ P S
Sbjct 104 DKASVTITSRYDGVIKRLYYNLDDIAYVGKPLIDIETEALKD--SEEDVVETPAVSHDEH 161
Query 145 PQQPSAPASSSRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYL 198
Q +G + LA+PAVRR A E + L V G+G G I KED+L++L
Sbjct 162 THQ------EIKGQKTLATPAVRRLAMENNIKLSEVVGSGKDGRILKEDILSFL 209
> dre:541388 dbt, im:7147214, zgc:103768; dihydrolipoamide branched
chain transacylase E2 (EC:2.3.1.168); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=493
Score = 128 bits (322), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 79/195 (40%), Positives = 104/195 (53%), Gaps = 9/195 (4%)
Query 6 KCSTWLSPAAAVSLRLLQEGLRSSPSTRGVFTVSHPRHGIVCFKLADIGEGIASVELTKW 65
C + L PAA + LR L + R T IV FKL+DIGEGI V + +W
Sbjct 25 NCCSKL-PAACLVLRPHSYSLVAGRQHRYFHTSYVAARPIVQFKLSDIGEGIMEVTVKEW 83
Query 66 YKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDIDSPDVE 125
Y K GD V + + +CEVQSDKA+V ITSRY G I KLY +G PL+DI++ +
Sbjct 84 YVKEGDKVSQFDSICEVQSDKASVTITSRYDGVIRKLYYDVDSIALVGKPLVDIETDGGQ 143
Query 126 ETQSQQPSPAAPPPS-EASKPQQPSAPASSSRGAEPLASPAVRRFAKEKGVNLDSVKGTG 184
Q+ P S E PQ+ +G + A+PAVRR A E + L V GTG
Sbjct 144 AESPQEDVVETPAVSQEEHSPQE-------IKGHKTQATPAVRRLAMENNIKLSEVVGTG 196
Query 185 ARGAITKEDVLNYLS 199
G I KED+LN+++
Sbjct 197 KDGRILKEDILNFIA 211
> xla:447616 dbt, MGC85493; dihydrolipoamide branched chain transacylase
E2 (EC:2.3.1.168); K09699 2-oxoisovalerate dehydrogenase
E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=492
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 69/167 (41%), Positives = 96/167 (57%), Gaps = 6/167 (3%)
Query 33 RGVFTVSHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEIT 92
R + T S IV FKL+DIGEGI V + WY K GD+V + + +CEVQSDKA+V IT
Sbjct 51 RSLRTASVLNGKIVQFKLSDIGEGITEVTVKDWYVKEGDSVSQFDSICEVQSDKASVTIT 110
Query 93 SRYSGKIVKLYAKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQPSAPA 152
SRY G I KL+ +T +G PL+DI++ +++ ++ P S Q
Sbjct 111 SRYDGVIRKLHYNVDETAYVGKPLVDIETDALKDVAPEEDVVETPAVSHDEHTHQ----- 165
Query 153 SSSRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYLS 199
+G + LA+PAVRR A E + L V G+G G I KED+L +L+
Sbjct 166 -EIKGHKTLATPAVRRLAMENNIKLSEVVGSGKDGRILKEDILGFLA 211
> tpv:TP04_0457 lipoamide transferase (EC:2.3.1.-); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=420
Score = 119 bits (298), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 62/110 (56%), Positives = 75/110 (68%), Gaps = 3/110 (2%)
Query 44 GIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLY 103
+ FKL+DIGEGI V+L KW K GD VEEME VC VQSDKAAVEITSRY+G + KLY
Sbjct 39 ALTTFKLSDIGEGINEVQLVKWEKNVGDEVEEMESVCTVQSDKAAVEITSRYTGVVKKLY 98
Query 104 AKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQPSAPAS 153
KEG+TVKIG PL+DID+ V+E P+ + ++ SK S P S
Sbjct 99 VKEGETVKIGGPLMDIDT--VDEVPDDTPNNISSNLND-SKRHYSSVPQS 145
> ath:AT3G06850 BCE2; BCE2; acetyltransferase/ alpha-ketoacid
dehydrogenase/ dihydrolipoamide branched chain acyltransferase
(EC:2.3.1.168); K09699 2-oxoisovalerate dehydrogenase E2
component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=483
Score = 113 bits (282), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 62/154 (40%), Positives = 87/154 (56%), Gaps = 8/154 (5%)
Query 44 GIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLY 103
G++ LA GEGIA EL KW+ K GD+VEE + +CEVQSDKA +EITSR+ GK+ +
Sbjct 74 GLIDVPLAQTGEGIAECELLKWFVKEGDSVEEFQPLCEVQSDKATIEITSRFKGKVALIS 133
Query 104 AKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLAS 163
GD +K+G L+ + D Q S SE + GA L++
Sbjct 134 HSPGDIIKVGETLVRLAVED------SQDSLLTTDSSEIVTLGGSKQGTENLLGA--LST 185
Query 164 PAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNY 197
PAVR AK+ G++++ + GTG G + KEDVL +
Sbjct 186 PAVRNLAKDLGIDINVITGTGKDGRVLKEDVLRF 219
> pfa:PFC0170c dihydrolipoamide acyltransferase, putative (EC:2.3.1.-);
K09699 2-oxoisovalerate dehydrogenase E2 component
(dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=448
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 66/163 (40%), Positives = 87/163 (53%), Gaps = 1/163 (0%)
Query 33 RGVFTVSHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEIT 92
R S IV KL DIGEGI+ VE+TKW+K GD V EME + VQSDKAAV+IT
Sbjct 21 RHYLNTSAIHFKIVKCKLFDIGEGISEVEITKWHKNEGDQVSEMESLLTVQSDKAAVDIT 80
Query 93 SRYSGKIVKLYAKEGDTVKIGAPLIDIDS-PDVEETQSQQPSPAAPPPSEASKPQQPSAP 151
S+Y+G +VK Y E D +K+G+ +ID+ D+ E ++ E S
Sbjct 81 SKYNGVLVKKYLNENDMLKVGSYFCEIDTDDDIIERDEEEVEKEENNKKEEDGESDLSLN 140
Query 152 ASSSRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDV 194
S ASP V+R AKE VNL+ V + I+ ED+
Sbjct 141 DDISNNDYIKASPGVKRKAKEYKVNLNKVGDYFNKVNISLEDL 183
> ath:AT3G13930 dihydrolipoamide S-acetyltransferase, putative
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=539
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 57/172 (33%), Positives = 86/172 (50%), Gaps = 35/172 (20%)
Query 61 ELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEG-DTVKIGAPLIDI 119
+ +W KK GD V E +CEV++DKA VE+ G + K+ +EG +++G +I I
Sbjct 127 NIARWLKKEGDKVAPGEVLCEVETDKATVEMECMEEGFLAKIVKEEGAKEIQVGE-VIAI 185
Query 120 ---DSPDVEETQSQQPS------------------------PAAPPPSEASKPQQPSAPA 152
D D+++ + PS PA+ P ++ SKP SAP+
Sbjct 186 TVEDEDDIQKFKDYTPSSDTGPAAPEAKPAPSLPKEEKVEKPASAPEAKISKPS--SAPS 243
Query 153 SSSRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYLSSGASE 204
A PLA R+ A++ V L S+KGTG G I K DV ++L+SG+ E
Sbjct 244 EDRIFASPLA----RKLAEDNNVPLSSIKGTGPEGRIVKADVEDFLASGSKE 291
> ath:AT1G54220 dihydrolipoamide S-acetyltransferase, putative
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=539
Score = 71.6 bits (174), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 50/162 (30%), Positives = 76/162 (46%), Gaps = 23/162 (14%)
Query 61 ELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDT-VKIGAPLIDI 119
+ +W KK GD V E +CEV++DKA VE+ G + K+ EG +++G +I I
Sbjct 127 NIARWLKKEGDKVAPGEVLCEVETDKATVEMECMEEGYLAKIVKAEGSKEIQVGE-VIAI 185
Query 120 DSPDVEET----------------QSQQPSPAAPPPSEASKPQQP-----SAPASSSRGA 158
D E+ +P+PA P + +P P S P++ G
Sbjct 186 TVEDEEDIGKFKDYTPSSTADAAPTKAEPTPAPPKEEKVKQPSSPPEPKASKPSTPPTGD 245
Query 159 EPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYLSS 200
ASP R+ A++ V L ++GTG G I K D+ YL+S
Sbjct 246 RVFASPLARKLAEDNNVPLSDIEGTGPEGRIVKADIDEYLAS 287
> xla:398314 dlat; dihydrolipoamide S-acetyltransferase (EC:2.3.1.12);
K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=628
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 54/180 (30%), Positives = 87/180 (48%), Gaps = 23/180 (12%)
Query 39 SHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGK 98
++P H +C + +V+ KW KK G+ + E + + E+++DKA + G
Sbjct 191 TYPNHMKICLPALSPTMTMGTVQ--KWEKKVGEKLSEGDLLAEIETDKATIGFEVPEEGY 248
Query 99 IVKLYAKEGD-TVKIGAPLI-------DIDS-PDVEETQSQ---QPSPAAPPPSEASKPQ 146
+ K+ EG V +G PL DI S D +E+ +P A P P+ AS P
Sbjct 249 LAKILVAEGTRDVPLGTPLCIIVEKESDISSFADYKESTGVVDIKPQHAPPTPTAASVPV 308
Query 147 QPSA-----PASSSRGAEP----LASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNY 197
P A P S+ + P SP ++ A EKG+++ VKG+G G ITK+D+ ++
Sbjct 309 PPVAVSTPAPTPSAAPSAPKGRVFVSPLAKKLAAEKGIDIKQVKGSGPEGRITKKDIDSF 368
Score = 38.1 bits (87), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEG 107
+ +W KK GD + E + + EV++DKA V S G + K+ EG
Sbjct 89 IARWEKKEGDKINEGDLIAEVETDKATVGFESLEEGYMAKILVAEG 134
> dre:393532 pdhx, MGC66110, zgc:66110, zgc:85977; pyruvate dehydrogenase
complex, component X; K13997 dihydrolipoamide dehydrogenase-binding
protein of pyruvate dehydrogenase complex
Length=490
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 57/198 (28%), Positives = 86/198 (43%), Gaps = 29/198 (14%)
Query 26 LRSSPSTRG--------VFTVSHPRHGIVCFK--LADIGEGIASVELTKWYKKTGDTVEE 75
LR SP T G ++ R G+ K + + + + KW KK G+ V
Sbjct 33 LRESPRTAGWSRSAARPLYQSCRARAGVCPLKVQMPALSPTMEEGNIVKWLKKEGEDVAA 92
Query 76 MEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDT-VKIGAPLIDIDSPDVEETQSQQPS- 133
+ +CE+++DKA V + S G + ++ +EG V++G + + S + Q + P+
Sbjct 93 GDALCEIETDKAVVVMESNEDGVLARILVQEGSRGVRLGTLIALMVSEGEDWKQVEIPAL 152
Query 134 ----------PAAPPPSEASKPQQPSAPASSSRGAEPL--ASPAVRRFAKEKGVNLDSVK 181
P A PP+ S P PA PL SPA R G++
Sbjct 153 EPVTPPTAALPTAAPPTAGSAP-----PALRQSVPTPLLRLSPAARHILDTHGLDPHQAT 207
Query 182 GTGARGAITKEDVLNYLS 199
+G RG ITKED LN LS
Sbjct 208 ASGPRGIITKEDALNLLS 225
> xla:447247 pdhx, MGC86218; pyruvate dehydrogenase complex, component
X; K13997 dihydrolipoamide dehydrogenase-binding protein
of pyruvate dehydrogenase complex
Length=478
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 70/140 (50%), Gaps = 3/140 (2%)
Query 61 ELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD-TVKIGAPLIDI 119
+ KW KK G++V + +CE+++DKA V + S G + K+ +EG V++G+ + +
Sbjct 59 NIVKWLKKEGESVSAGDALCEIETDKAVVTMESNDDGVLAKILVEEGSKNVRLGSLIALL 118
Query 120 DSPDVEETQSQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLASPAVRRFAKEKGVNLDS 179
+ Q PS P + A+ + + + RG SPA R G++ S
Sbjct 119 VEEGQDWKQVHVPSVKVSPTTVAAATKIANVAPVAKRGLR--MSPAARHIIDTHGLDTGS 176
Query 180 VKGTGARGAITKEDVLNYLS 199
+ +G RG ITKED L L+
Sbjct 177 ITPSGPRGIITKEDALKCLA 196
> sce:YNL071W LAT1, ODP2, PDA2; Dihydrolipoamide acetyltransferase
component (E2) of pyruvate dehydrogenase complex, which
catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2 component
(dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=482
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 51/185 (27%), Positives = 83/185 (44%), Gaps = 27/185 (14%)
Query 39 SHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGK 98
S+P H I+ + + + L W KK GD + E + E+++DKA ++ + G
Sbjct 30 SYPEHTIIG--MPALSPTMTQGNLAAWTKKEGDQLSPGEVIAEIETDKAQMDFEFQEDGY 87
Query 99 IVKLYAKEG-DTVKIGAPLIDI--DSPDV--------EETQSQQPSPAAPPPSEASKPQQ 147
+ K+ EG + + P+ D DV E++ S + P+E ++
Sbjct 88 LAKILVPEGTKDIPVNKPIAVYVEDKADVPAFKDFKLEDSGSDSKTSTKAQPAEPQAEKK 147
Query 148 PSAPASSSRGAEP--------------LASPAVRRFAKEKGVNLDSVKGTGARGAITKED 193
APA ++ + P ASP + A EKG++L V GTG RG ITK D
Sbjct 148 QEAPAEETKTSAPEAKKSDVAAPQGRIFASPLAKTIALEKGISLKDVHGTGPRGRITKAD 207
Query 194 VLNYL 198
+ +YL
Sbjct 208 IESYL 212
> eco:b0727 sucB, ECK0715, JW0716; dihydrolipoyltranssuccinase
(EC:2.3.1.61); K00658 2-oxoglutarate dehydrogenase E2 component
(dihydrolipoamide succinyltransferase) [EC:2.3.1.61]
Length=405
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 72/148 (48%), Gaps = 6/148 (4%)
Query 52 DIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVK 111
D+ E +A + W+KK GD V E + E+++DK +E+ + G + + EG TV
Sbjct 10 DLPESVADATVATWHKKPGDAVVRDEVLVEIETDKVVLEVPASADGILDAVLEDEGTTVT 69
Query 112 IGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLASPAVRRFAK 171
L + E S +A +AS P Q + + + L SPA+RR
Sbjct 70 SRQIL-----GRLREGNSAGKETSAKSEEKASTPAQRQQASLEEQNNDAL-SPAIRRLLA 123
Query 172 EKGVNLDSVKGTGARGAITKEDVLNYLS 199
E ++ ++KGTG G +T+EDV +L+
Sbjct 124 EHNLDASAIKGTGVGGRLTREDVEKHLA 151
> mmu:235339 Dlat, 6332404G05Rik, DLTA, PDC-E2; dihydrolipoamide
S-acetyltransferase (E2 component of pyruvate dehydrogenase
complex) (EC:2.3.1.12); K00627 pyruvate dehydrogenase E2
component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=642
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 48/162 (29%), Positives = 76/162 (46%), Gaps = 28/162 (17%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDT-VKIGAPLIDID 120
+ +W KK G+ + E + + E+++DKA + + G + K+ EG V +GAPL I
Sbjct 234 VQRWEKKVGEKLSEGDLLAEIETDKATIGFEVQEEGYLAKILVPEGTRDVPLGAPLCII- 292
Query 121 SPDVEETQSQQPSPAAPPPSEASKPQ------------------QPSAPASSSRGAEP-- 160
E Q + A P+E + + QP AP S+ A P
Sbjct 293 ----VEKQEDIAAFADYRPTEVTSLKPQAAPPAPPPVAAVPPTPQPVAPTPSAAPAGPKG 348
Query 161 --LASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYLSS 200
SP ++ A EKG++L VKGTG G I K+D+ +++ S
Sbjct 349 RVFVSPLAKKLAAEKGIDLTQVKGTGPEGRIIKKDIDSFVPS 390
Score = 35.4 bits (80), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 56/119 (47%), Gaps = 8/119 (6%)
Query 10 WLSPAAAVSLRLLQEGLRSSPSTRGVFTVSHPRHGIVCFKLADIGEGIASVELTKWYKKT 69
W S + V L L SPS R + S P H V L + + + + +W KK
Sbjct 60 WSSGSGTVPRNRLLRQLLGSPSRR---SYSLPPHQKV--PLPSLSPTMQAGTIARWEKKE 114
Query 70 GDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD-TVKIGAPL-IDIDSP-DVE 125
G+ + E + + EV++DKA V S + K+ EG V +G+ + I ++ P D+E
Sbjct 115 GEKISEGDLIAEVETDKATVGFESLEECYMAKILVPEGTRDVPVGSIICITVEKPQDIE 173
> tpv:TP01_0262 dihydrolipoamide succinyltransferase; K00658 2-oxoglutarate
dehydrogenase E2 component (dihydrolipoamide succinyltransferase)
[EC:2.3.1.61]
Length=456
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 53/90 (58%), Gaps = 7/90 (7%)
Query 53 IGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKI 112
+G+ I+ LTKW GD + + + V++DK +V++ S +SG + K ++ GDT+ +
Sbjct 80 LGDSISEGTLTKWAVSVGDYLNVDDLIAVVETDKVSVDVNSPFSGVLTKTFSNTGDTILV 139
Query 113 GAPLIDI-------DSPDVEETQSQQPSPA 135
G PL++I D P ++T+ + P+PA
Sbjct 140 GKPLVEIDLAGKPSDKPPEKKTEDKPPTPA 169
> eco:b0115 aceF, ECK0114, JW0111; pyruvate dehydrogenase, dihydrolipoyltransacetylase
component E2 (EC:2.3.1.12); K00627
pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase)
[EC:2.3.1.12]
Length=630
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 48/147 (32%), Positives = 75/147 (51%), Gaps = 8/147 (5%)
Query 60 VELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDI 119
VE+T+ K GD V + + V+ DKA++E+ + ++G + +L GD VK G+ ++
Sbjct 219 VEVTEVMVKVGDKVAAEQSLITVEGDKASMEVPAPFAGVVKELKVNVGDKVKTGSLIMIF 278
Query 120 DSPDVEETQSQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLA--------SPAVRRFAK 171
+ + AA P A +APA+ + G A +P +RR A+
Sbjct 279 EVEGAAPAAAPAKQEAAAPAPAAKAEAPAAAPAAKAEGKSEFAENDAYVHATPLIRRLAR 338
Query 172 EKGVNLDSVKGTGARGAITKEDVLNYL 198
E GVNL VKGTG +G I +EDV Y+
Sbjct 339 EFGVNLAKVKGTGRKGRILREDVQAYV 365
Score = 36.6 bits (83), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 38/66 (57%), Gaps = 2/66 (3%)
Query 52 DIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVK 111
DIG VE+T+ K GD VE + + V+ DKA++E+ + ++G + ++ GD V
Sbjct 112 DIGSD--EVEVTEILVKVGDKVEAEQSLITVEGDKASMEVPAPFAGTVKEIKVNVGDKVS 169
Query 112 IGAPLI 117
G+ ++
Sbjct 170 TGSLIM 175
Score = 36.2 bits (82), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query 48 FKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEG 107
K+ DIG VE+T+ K GD VE + + V+ DKA++E+ S +G + ++ G
Sbjct 5 IKVPDIGAD--EVEITEILVKVGDKVEAEQSLITVEGDKASMEVPSPQAGIVKEIKVSVG 62
Query 108 DTVKIGAPLI 117
D + GA ++
Sbjct 63 DKTQTGALIM 72
> bbo:BBOV_IV004840 23.m06243; dihydrolipoamide succinyltransferase
(EC:2.3.1.61); K00658 2-oxoglutarate dehydrogenase E2
component (dihydrolipoamide succinyltransferase) [EC:2.3.1.61]
Length=402
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 43/142 (30%), Positives = 69/142 (48%), Gaps = 11/142 (7%)
Query 48 FKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEG 107
KL +G+ I+ L++W K G++VE E + V++DK V+I S SG IVK + +
Sbjct 58 MKLPSLGDSISEGTLSEWKKNVGESVEVDEPIAIVETDKVTVDINSTLSGVIVKQHYEVD 117
Query 108 DTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLASPAVR 167
DTV +G P ID+D+ S AAP + + + P + + EP +P
Sbjct 118 DTVLVGKPFIDVDAGG---------SAAAPAETASGVDSKSPEPVAEVKADEP--APTET 166
Query 168 RFAKEKGVNLDSVKGTGARGAI 189
R + ++ V+ T R I
Sbjct 167 RVCYQLSLHNVQVQMTRMRKRI 188
> hsa:1737 DLAT, DLTA, PDC-E2, PDCE2; dihydrolipoamide S-acetyltransferase
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2
component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=647
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 52/190 (27%), Positives = 82/190 (43%), Gaps = 36/190 (18%)
Query 39 SHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGK 98
S+P H V + +V+ +W KK G+ + E + + E+++DKA + + G
Sbjct 214 SYPPHMQVLLPALSPTMTMGTVQ--RWEKKVGEKLSEGDLLAEIETDKATIGFEVQEEGY 271
Query 99 IVKLYAKEGD-TVKIGAPLIDI--DSPDVEETQSQQPS---------------------- 133
+ K+ EG V +G PL I D+ +P+
Sbjct 272 LAKILVPEGTRDVPLGTPLCIIVEKEADISAFADYRPTEVTDLKPQVPPPTPPPVAAVPP 331
Query 134 ---PAAPPPSEASKPQQPSAPASSSRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAIT 190
P AP PS A P P+ P SP ++ A EKG++L VKGTG G IT
Sbjct 332 TPQPLAPTPS-APCPATPAGPKGRV-----FVSPLAKKLAVEKGIDLTQVKGTGPDGRIT 385
Query 191 KEDVLNYLSS 200
K+D+ +++ S
Sbjct 386 KKDIDSFVPS 395
Score = 35.8 bits (81), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD-TVKIGAPL-IDI 119
+ +W KK GD + E + + EV++DKA V S + K+ EG V IGA + I +
Sbjct 108 IARWEKKEGDKINEGDLIAEVETDKATVGFESLEECYMAKILVAEGTRDVPIGAIICITV 167
Query 120 DSP-DVE 125
P D+E
Sbjct 168 GKPEDIE 174
> tgo:TGME49_085680 dihydrolipoamide acyltransferase, putative
(EC:2.4.1.115 2.3.1.61)
Length=470
Score = 59.3 bits (142), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 48 FKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEG 107
K+ +G+ I L +W KK GD V E +C +++DK VEI S SG ++ A+EG
Sbjct 235 IKVPSLGDSITEGGLLEWRKKVGDFVLVDEVLCVIETDKVTVEIHSDCSGILLAQAAQEG 294
Query 108 DTVKIGAPLIDIDSPD 123
DTV++G+ L +D D
Sbjct 295 DTVQVGSQLAVLDYSD 310
> tgo:TGME49_019550 dihydrolipoamide succinyltransferase component
of 2-oxoglutaratedehydrogenase complex, putative (EC:2.3.1.61);
K00658 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=470
Score = 59.3 bits (142), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 56/109 (51%), Gaps = 20/109 (18%)
Query 53 IGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKI 112
+G+ I L +W K+ G+ V+E E V + +DK +V+I + +G+IV+ A GDTV++
Sbjct 101 MGDSITEGSLNEWKKQPGEYVKEGELVAVIDTDKVSVDINAPQAGRIVRFEANAGDTVEV 160
Query 113 GAPLIDIDSPDVEETQSQQPSPA-------------APPPSEASKPQQP 148
G PL ID + QP PA P +EA+KP P
Sbjct 161 GKPLYVIDP-------TAQPDPAELAAAAAAAAAPATPVKTEAAKPVSP 202
> pfa:PF13_0121 dihydrolipamide succinyltransferase component
of 2-oxoglutarate dehydrogenase complex (EC:2.3.1.61); K00658
2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=421
Score = 56.2 bits (134), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 45/73 (61%), Gaps = 0/73 (0%)
Query 49 KLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD 108
K+ +G+ I + +W KK GD V+ E + + +DK +V+I S+ SG + K++A GD
Sbjct 49 KVPRLGDSITEGTINEWKKKVGDYVKADETITIIDTDKVSVDINSKVSGGLSKIFADVGD 108
Query 109 TVKIGAPLIDIDS 121
V + APL +ID+
Sbjct 109 VVLVDAPLCEIDT 121
> bbo:BBOV_III003270 17.m07312; biotin-requiring enzyme family
protein
Length=177
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 44/74 (59%), Gaps = 0/74 (0%)
Query 49 KLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD 108
K+ IG + ++ +W+K+ GD V+ + +C +++D+ V + S+ SG IV+ EG
Sbjct 85 KVPHIGRDVKHSKIQQWHKQRGDEVDVGDLICVLETDQVLVNVQSQLSGTIVETVGNEGC 144
Query 109 TVKIGAPLIDIDSP 122
VK+GA LI I P
Sbjct 145 RVKVGADLIIIRRP 158
> dre:324201 dlat, wu:fc14f10, wu:fc21f08, wu:fc86g11, wu:fj57d06;
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate
dehydrogenase complex) (EC:2.3.1.12); K00627 pyruvate
dehydrogenase E2 component (dihydrolipoamide acetyltransferase)
[EC:2.3.1.12]
Length=652
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 41/155 (26%), Positives = 73/155 (47%), Gaps = 20/155 (12%)
Query 64 KWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD-TVKIGAPLIDIDSP 122
+W KK G+ + E + + E+++DKA + + G + K+ EG V +G PL I
Sbjct 237 RWEKKVGEKLSEGDLLAEIETDKATIGFEVQEEGYLAKIMISEGTRDVPLGTPLCIIVEK 296
Query 123 DVEET----QSQQPSPAAPPPS---------------EASKPQQPSAPASSSRGAEPLAS 163
+ + + + A+PPP+ P A +++R AS
Sbjct 297 ESDISAFADYVETGVAASPPPAPTLVATPPPAAAPAAPIPAPAAAPAAPAAARKGRVFAS 356
Query 164 PAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYL 198
P ++ A EKGV++ V GTG G +TK+D+ +++
Sbjct 357 PLAKKLAAEKGVDITQVTGTGPDGRVTKKDIDSFV 391
Score = 37.0 bits (84), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD-TVKIGAPL-IDI 119
+ +W KK GD + E + + EV++DKA V + K+ EG V IGA + I +
Sbjct 109 IARWEKKEGDKINEGDLIAEVETDKATVGFEMLEECYLAKILVAEGTRDVPIGAVICITV 168
Query 120 DSPDV 124
D P++
Sbjct 169 DKPEL 173
> ath:AT3G52200 LTA3; LTA3; ATP binding / dihydrolipoyllysine-residue
acetyltransferase; K00627 pyruvate dehydrogenase E2
component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=637
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 41/143 (28%), Positives = 67/143 (46%), Gaps = 16/143 (11%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD-TVKIGAP--LID 118
+ KW+KK GD +E + + E+++DKA +E S G + K+ EG V +G P LI
Sbjct 229 IAKWWKKEGDKIEVGDVIGEIETDKATLEFESLEEGYLAKILIPEGSKDVAVGKPIALIV 288
Query 119 IDSPDVEETQ------SQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLASPAVRRFAKE 172
D+ +E + S+ + P S KP + A + SPA + E
Sbjct 289 EDAESIEAIKSSSAGSSEVDTVKEVPDSVVDKPTERKAGFTK-------ISPAAKLLILE 341
Query 173 KGVNLDSVKGTGARGAITKEDVL 195
G+ S++ +G G + K DV+
Sbjct 342 HGLEASSIEASGPYGTLLKSDVV 364
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEG 107
+ KW KK GD VE + +CE+++DKA VE S+ G + K+ EG
Sbjct 102 VVKWMKKEGDKVEVGDVLCEIETDKATVEFESQEEGFLAKILVTEG 147
> hsa:8050 PDHX, DLDBP, E3BP, OPDX, PDX1, proX; pyruvate dehydrogenase
complex, component X; K13997 dihydrolipoamide dehydrogenase-binding
protein of pyruvate dehydrogenase complex
Length=486
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 40/149 (26%), Positives = 64/149 (42%), Gaps = 11/149 (7%)
Query 61 ELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGD-TVKIGAPLIDI 119
+ KW KK G+ V + +CE+++DKA V + + G + K+ +EG +++G+ + I
Sbjct 57 NIVKWLKKEGEAVSAGDALCEIETDKAVVTLDASDDGILAKIVVEEGSKNIRLGSLIGLI 116
Query 120 DSPDVEETQSQQPSPAAPPP-------SEASKPQQPSAPASSSRGAEPL---ASPAVRRF 169
+ + P PPP S Q S P L SPA R
Sbjct 117 VEEGEDWKHVEIPKDVGPPPPVSKPSEPRPSPEPQISIPVKKEHIPGTLRFRLSPAARNI 176
Query 170 AKEKGVNLDSVKGTGARGAITKEDVLNYL 198
++ ++ TG RG TKED L +
Sbjct 177 LEKHSLDASQGTATGPRGIFTKEDALKLV 205
> ath:AT4G26910 2-oxoacid dehydrogenase family protein (EC:2.3.1.61);
K00658 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=464
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 54/112 (48%), Gaps = 14/112 (12%)
Query 53 IGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKI 112
+GE I L + KK G+ V+ E + ++++DK ++I S SG I + EGDTV+
Sbjct 100 MGESITDGTLATFLKKPGERVQADEAIAQIETDKVTIDIASPASGVIQEFLVNEGDTVEP 159
Query 113 GAPLIDID-----------SPDVEETQSQQPSPAAPPPSEASKPQQPSAPAS 153
G + I S + ET +PS PP + KP+ SAP +
Sbjct 160 GTKVAIISKSEDTASQVTPSQKIPETTDTKPS---PPAEDKQKPRVESAPVA 208
> mmu:27402 Pdhx, AI481367, E3bp, Pdx1; pyruvate dehydrogenase
complex, component X; K13997 dihydrolipoamide dehydrogenase-binding
protein of pyruvate dehydrogenase complex
Length=501
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 43/159 (27%), Positives = 68/159 (42%), Gaps = 31/159 (19%)
Query 61 ELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEG-DTVKIGAPLIDI 119
+ KW +K G+ V + +CE+++DKA V + + G + K+ +EG +++G+ LI +
Sbjct 72 NIVKWLRKEGEAVSAGDSLCEIETDKAVVTLDANDDGILAKIVVEEGAKNIQLGS-LIAL 130
Query 120 ------DSPDVE-------------ETQSQQPSPAAPPPSEASKPQQPSAPASSSRG-AE 159
D VE QPSP +PQ P +G A
Sbjct 131 MVEEGEDWKQVEIPKDVSAPPPVSKPPAPTQPSP---------QPQIPCPARKEHKGTAR 181
Query 160 PLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYL 198
SPA R ++ ++ TG RG TKED L +
Sbjct 182 FRLSPAARNILEKHSLDASQGTATGPRGIFTKEDALKLV 220
> sce:YDR148C KGD2; Dihydrolipoyl transsuccinylase, component
of the mitochondrial alpha-ketoglutarate dehydrogenase complex,
which catalyzes the oxidative decarboxylation of alpha-ketoglutarate
to succinyl-CoA in the TCA cycle; phosphorylated
(EC:2.3.1.61); K00658 2-oxoglutarate dehydrogenase E2 component
(dihydrolipoamide succinyltransferase) [EC:2.3.1.61]
Length=463
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 56/112 (50%), Gaps = 3/112 (2%)
Query 36 FTVSHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRY 95
F+++ R ++ + E + L ++ K GD ++E E + +++DK +E+ S
Sbjct 64 FSITSNRFKSTSIEVPPMAESLTEGSLKEYTKNVGDFIKEDELLATIETDKIDIEVNSPV 123
Query 96 SGKIVKLYAKEGDTVKIGAPLIDIDSPDVEETQSQQPSPAAPPPSEASKPQQ 147
SG + KL K DTV +G L ++ E ++ + P P+E ++P Q
Sbjct 124 SGTVTKLNFKPEDTVTVGEELAQVEPG---EAPAEGSGESKPEPTEQAEPSQ 172
> ath:AT5G55070 2-oxoacid dehydrogenase family protein (EC:2.3.1.61);
K00658 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=464
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 36/61 (59%), Gaps = 0/61 (0%)
Query 53 IGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKI 112
+GE I L + KK GD VE E + ++++DK ++I S SG I + KEGDTV+
Sbjct 101 MGESITDGTLAAFLKKPGDRVEADEAIAQIETDKVTIDIASPASGVIQEFLVKEGDTVEP 160
Query 113 G 113
G
Sbjct 161 G 161
> hsa:1743 DLST, DLTS; dihydrolipoamide S-succinyltransferase
(E2 component of 2-oxo-glutarate complex) (EC:2.3.1.61); K00658
2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=453
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 46/91 (50%), Gaps = 7/91 (7%)
Query 35 VFTVSHPRHGIVC------FKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAA 88
VF+V R VC K E + ++ +W K GDTV E E VCE+++DK +
Sbjct 54 VFSVRFFRTTAVCKDDLVTVKTPAFAESVTEGDV-RWEKAVGDTVAEDEVVCEIETDKTS 112
Query 89 VEITSRYSGKIVKLYAKEGDTVKIGAPLIDI 119
V++ S +G I L +G V+ G PL +
Sbjct 113 VQVPSPANGVIEALLVPDGGKVEGGTPLFTL 143
> xla:380395 dlst, MGC53142; dihydrolipoamide S-succinyltransferase
(E2 component of 2-oxo-glutarate complex) (EC:2.3.1.61);
K00658 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=452
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 33/54 (61%), Gaps = 0/54 (0%)
Query 64 KWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLI 117
+W K GDTV E E VCE+++DK +V++ S +G I L +G V+ G PL
Sbjct 86 RWEKAVGDTVSEDEVVCEIETDKTSVQVPSPSAGVIEALLVPDGGKVEGGTPLF 139
> ath:AT1G34430 EMB3003 (embryo defective 3003); acyltransferase/
dihydrolipoyllysine-residue acetyltransferase/ protein binding;
K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=465
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 41/164 (25%), Positives = 66/164 (40%), Gaps = 31/164 (18%)
Query 61 ELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDID 120
++ W K GD + + E V V+SDKA +++ + Y G + + +EG +G+ + +
Sbjct 55 KIVSWVKSEGDKLNKGESVVVVESDKADMDVETFYDGYLAAIMVEEGGVAPVGSAIALLA 114
Query 121 SPD------------------------------VEETQSQQPSPAAPPPSEASKPQQPSA 150
+ VE S + AA P S +
Sbjct 115 ETEDEIADAKAKASGGGGGGDSKAPPASPPTAAVEAPVSVEKKVAAAPVSIKAVAASAVH 174
Query 151 PASSSRGAEPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDV 194
PAS G +ASP ++ AKE V L + G+G G I +DV
Sbjct 175 PASEG-GKRIVASPYAKKLAKELKVELAGLVGSGPMGRIVAKDV 217
> dre:368262 dlst, cb60, wu:fc39d01; dihydrolipoamide S-succinyltransferase
(EC:2.3.1.61); K00658 2-oxoglutarate dehydrogenase
E2 component (dihydrolipoamide succinyltransferase) [EC:2.3.1.61]
Length=458
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 47/83 (56%), Gaps = 2/83 (2%)
Query 37 TVSHPRHGIVCFKLADIGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYS 96
T +H R+ ++ K E + ++ +W K GD+V E E VCE+++DK +V++ S +
Sbjct 63 TAAH-RNEVITVKTPAFAESVTEGDV-RWEKAVGDSVAEDEVVCEIETDKTSVQVPSPAA 120
Query 97 GKIVKLYAKEGDTVKIGAPLIDI 119
G I +L +G V+ G PL +
Sbjct 121 GVIEELLVPDGGKVEGGTPLFKL 143
> mmu:78920 Dlst, 1600017E01Rik, 4632413C10Rik, 4930529O08Rik,
DLTS; dihydrolipoamide S-succinyltransferase (E2 component
of 2-oxo-glutarate complex) (EC:2.3.1.61); K00658 2-oxoglutarate
dehydrogenase E2 component (dihydrolipoamide succinyltransferase)
[EC:2.3.1.61]
Length=454
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 63 TKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDI 119
+W K GD V E E VCE+++DK +V++ S +G I L +G V+ G PL +
Sbjct 88 VRWEKAVGDAVAEDEVVCEIETDKTSVQVPSPANGIIEALLVPDGGKVEGGTPLFTL 144
> cel:F23B12.5 hypothetical protein; K00627 pyruvate dehydrogenase
E2 component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=507
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 41/144 (28%), Positives = 61/144 (42%), Gaps = 28/144 (19%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDT-VKIGAPLIDI- 119
+ W KK GD + E + +CE+++DKA + + G + K+ +EG V IG L I
Sbjct 94 VVSWQKKEGDQLSEGDLLCEIETDKATMGFETPEEGYLAKILIQEGSKDVPIGKLLCIIV 153
Query 120 -DSPDVEETQS----------------QQPSPAAPPPSEASKP-----QQPSAPASS--- 154
+ DV + + P PA P S P Q PS P S+
Sbjct 154 DNEADVAAFKDFKDDGASSGGSAPAAEKAPEPAKPAASSQPSPPAQMYQAPSVPKSAPIP 213
Query 155 -SRGAEPLASPAVRRFAKEKGVNL 177
S ASP ++ A E G++L
Sbjct 214 HSSSGRVSASPFAKKLAAENGLDL 237
> ath:AT3G25860 LTA2; LTA2; dihydrolipoyllysine-residue acetyltransferase
Length=480
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 50/193 (25%), Positives = 77/193 (39%), Gaps = 27/193 (13%)
Query 29 SPSTRGVF------TVSHPRHGIVCFKLADIGEGIASVELTK-----WYKKTGDTVEEME 77
SPS R V SH R V K+ +I S +T+ W K G+ + + E
Sbjct 28 SPSLRSVVFRSTTPATSHRRSMTVRSKIREIFMPALSSTMTEGKIVSWIKTEGEKLAKGE 87
Query 78 EVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPL----------------IDIDS 121
V V+SDKA +++ + Y G + + EG+T +GA + S
Sbjct 88 SVVVVESDKADMDVETFYDGYLAAIVVGEGETAPVGAAIGLLAETEAEIEEAKSKAASKS 147
Query 122 PDVEETQSQQPSPAAPPPSEASKPQQPSAPASSSRGAEPLASPAVRRFAKEKGVNLDSVK 181
P + Q A S + +A+P ++ AK+ V+++SV
Sbjct 148 SSSVAEAVVPSPPPVTSSPAPAIAQPAPVTAVSDGPRKTVATPYAKKLAKQHKVDIESVA 207
Query 182 GTGARGAITKEDV 194
GTG G IT DV
Sbjct 208 GTGPFGRITASDV 220
> cel:W02F12.5 hypothetical protein; K00658 2-oxoglutarate dehydrogenase
E2 component (dihydrolipoamide succinyltransferase)
[EC:2.3.1.61]
Length=463
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Query 64 KWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDIDSPD 123
+W K+ GD V E E V E+++DK +VE+ + +G IV+ ++G V L + P
Sbjct 81 RWLKQKGDHVNEDELVAEIETDKTSVEVPAPQAGTIVEFLVEDGAKVTAKQKLYKL-QPG 139
Query 124 VEETQSQQPS---PAAPPPSEASKP 145
S P+ P + P E SKP
Sbjct 140 AGGGSSSAPAKEEPKSAPAKEESKP 164
> dre:100330874 pyruvate dehydrogenase complex, component X-like
Length=181
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 47/99 (47%), Gaps = 11/99 (11%)
Query 26 LRSSPSTRG--------VFTVSHPRHGIVCFK--LADIGEGIASVELTKWYKKTGDTVEE 75
LR SP T G ++ R G+ K + + + + KW KK G+ V
Sbjct 33 LRESPRTAGWSRSAARPLYQSCRARAGVCPLKVQMPALSPTMEEGNIVKWLKKEGEDVAA 92
Query 76 MEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDT-VKIG 113
+ +CE+++DKA V + S G + ++ +EG V++G
Sbjct 93 GDALCEIETDKAVVVMESNEDGVLARILVQEGSRGVRLG 131
> tpv:TP03_0320 hypothetical protein
Length=267
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 53 IGEGIASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKI 112
+G I ++ KW KK GD V + +C +++D ++ S+ +G I + +EG V
Sbjct 106 LGRNITRCKIYKWLKKPGDYVNLNDLLCIIETDLIFGKVYSKVTGTIFESVHQEGSMVNC 165
Query 113 GAPLI 117
G+ L+
Sbjct 166 GSDLM 170
> pfa:PF10_0407 dihydrolipoamide acyltransferase, putative (EC:2.3.1.12);
K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=640
Score = 40.4 bits (93), Expect = 0.005, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 3/71 (4%)
Query 65 WYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDI---DS 121
W K D V++ + + V+ DK+ +E+ S YSG I KL KEG V + + I +
Sbjct 202 WLKNENDFVKKNDLLLYVEDDKSTIEVESPYSGIIKKLLVKEGQFVDLDKEVAIISITEE 261
Query 122 PDVEETQSQQP 132
D E+ + ++P
Sbjct 262 KDNEKEKIEEP 272
> tgo:TGME49_006610 biotin requiring domain-containing protein
/ 2-oxo acid dehydrogenases acyltransferase catalytic domain-containing
protein (EC:6.4.1.4 2.3.1.12 2.3.1.168); K00627
pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase)
[EC:2.3.1.12]
Length=932
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 57 IASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGA 114
+ + +T W KK G+ V + + + V+SDKA +++ + + G + + +EG TV +G+
Sbjct 252 LKTARMTVWRKKEGEKVNKGDVLFVVESDKADMDVEAPHDGVLAHIAVREGVTVDVGS 309
Score = 36.6 bits (83), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 61 ELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIG 113
++ W K+ GD VE + + V+SDKA +++ + SG + +EGD +G
Sbjct 150 KVVTWSKQVGDRVEPGDVLMVVESDKADMDVEAFDSGFMAMHLVREGDAAPVG 202
Score = 35.8 bits (81), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 57 IASVELTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIG 113
+ S +++KW K GD V + + V+SDKA +++ S G + + EG++ +G
Sbjct 358 MTSGKVSKWNKAVGDAVHVGDTLMVVESDKADMDVESFDEGYLAAITVAEGESAPVG 414
Score = 33.5 bits (75), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 159 EPLASPAVRRFAKEKGVNLDSVKGTGARGAITKEDVLNYL 198
+PLA+ AK+ +NL+ VKGTG IT DV +L
Sbjct 611 QPLATFNAIELAKKNKLNLEEVKGTGTNRRITAADVRQHL 650
> sce:YGR193C PDX1; Dihydrolipoamide dehydrogenase (E3)-binding
protein (E3BP) of the mitochondrial pyruvate dehydrogenase
(PDH) complex, plays a structural role in the complex by binding
and positioning E3 to the dihydrolipoamide acetyltransferase
(E2) core
Length=410
Score = 38.1 bits (87), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 36/162 (22%), Positives = 68/162 (41%), Gaps = 25/162 (15%)
Query 62 LTKWYKKTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDT-VKIGAPLIDID 120
+ W K G+ + + EV++DK+ +++ + GK+ K+ EG V +G P+ I
Sbjct 49 IVSWKYKVGEPFSAGDVILEVETDKSQIDVEALDDGKLAKILKDEGSKDVDVGEPIAYIA 108
Query 121 SPDVEETQSQQPSPAAPPPSEASKPQQPSAPAS--------------------SSRGAEP 160
D + + P A +++ + ++PSA ++ S E
Sbjct 109 DVDDDLATIKLPQEANTANAKSIEIKKPSADSTEATQQHLKKATVTPIKTVDGSQANLEQ 168
Query 161 LASPAVRRFAKEKGVN----LDSVKGTGARGAITKEDVLNYL 198
P+V E ++ L + +G+ G + K DVL YL
Sbjct 169 TLLPSVSLLLAENNISKQKALKEIAPSGSNGRLLKGDVLAYL 210
> dre:437019 pcca, wu:fb92g02, zgc:100925; propionyl-Coenzyme
A carboxylase, alpha polypeptide (EC:6.4.1.3); K01965 propionyl-CoA
carboxylase alpha chain [EC:6.4.1.3]
Length=709
Score = 37.0 bits (84), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 33/53 (62%), Gaps = 0/53 (0%)
Query 68 KTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDID 120
K GDTV E +E+C +++ K +T+ + K+ ++ K GDTV G L++++
Sbjct 657 KPGDTVAEGQEICVIEAMKMQNSMTAAKTAKVKSVHCKAGDTVGEGDLLVELE 709
> mmu:110821 Pcca, C79630; propionyl-Coenzyme A carboxylase, alpha
polypeptide (EC:6.4.1.3); K01965 propionyl-CoA carboxylase
alpha chain [EC:6.4.1.3]
Length=724
Score = 35.8 bits (81), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 32/53 (60%), Gaps = 0/53 (0%)
Query 68 KTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDID 120
K GD V E +E+C +++ K +T+ GK+ ++ K GDTV G L++++
Sbjct 672 KPGDMVAEGQEICVIEAMKMQNSMTAGKMGKVKLVHCKAGDTVGEGDLLVELE 724
> ath:AT3G56130 biotin/lipoyl attachment domain-containing protein
Length=194
Score = 34.3 bits (77), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 15/52 (28%), Positives = 31/52 (59%), Gaps = 0/52 (0%)
Query 68 KTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDI 119
K GD ++E + + + + +TS +G+++KL + +GD+V G PL+ +
Sbjct 133 KEGDAIKEGQVIGYLHQLGTELPVTSDVAGEVLKLLSDDGDSVGYGDPLVAV 184
> hsa:5095 PCCA; propionyl CoA carboxylase, alpha polypeptide
(EC:6.4.1.3); K01965 propionyl-CoA carboxylase alpha chain [EC:6.4.1.3]
Length=728
Score = 33.5 bits (75), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 32/53 (60%), Gaps = 0/53 (0%)
Query 68 KTGDTVEEMEEVCEVQSDKAAVEITSRYSGKIVKLYAKEGDTVKIGAPLIDID 120
K GD V E +E+C +++ K +T+ +G + ++ + GDTV G L++++
Sbjct 676 KPGDAVAEGQEICVIEAMKMQNSMTAGKTGTVKSVHCQAGDTVGEGDLLVELE 728
Lambda K H
0.308 0.126 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6253620652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40