bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3566_orf2
Length=251
Score E
Sequences producing significant alignments: (Bits) Value
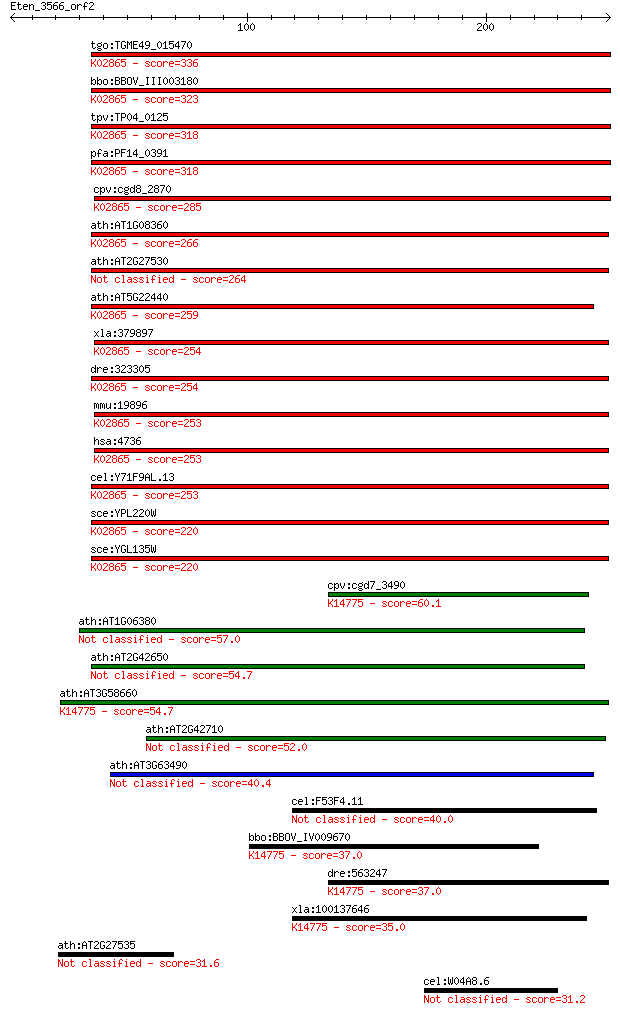
tgo:TGME49_015470 60S ribosomal protein L10a, putative ; K0286... 336 4e-92
bbo:BBOV_III003180 17.m07305; ribosomal protein L1; K02865 lar... 323 4e-88
tpv:TP04_0125 60S ribosomal protein L1; K02865 large subunit r... 318 1e-86
pfa:PF14_0391 60S ribosomal protein L1, putative; K02865 large... 318 1e-86
cpv:cgd8_2870 60S ribosomal protein L10A ; K02865 large subuni... 285 1e-76
ath:AT1G08360 60S ribosomal protein L10A (RPL10aA); K02865 lar... 266 7e-71
ath:AT2G27530 PGY1; PGY1 (PIGGYBACK1); RNA binding / structura... 264 2e-70
ath:AT5G22440 60S ribosomal protein L10A (RPL10aC); K02865 lar... 259 9e-69
xla:379897 rpl10a, MGC53614; ribosomal protein L10a (EC:3.6.5.... 254 2e-67
dre:323305 rpl10a, MGC86881, wu:fb94f08, zgc:73082, zgc:86881;... 254 2e-67
mmu:19896 Rpl10a, CsA-19, Nedd6; ribosomal protein L10A (EC:3.... 253 3e-67
hsa:4736 RPL10A, Csa-19, NEDD6; ribosomal protein L10a; K02865... 253 3e-67
cel:Y71F9AL.13 rpl-1; Ribosomal Protein, Large subunit family ... 253 4e-67
sce:YPL220W RPL1A, SSM1; L1A; K02865 large subunit ribosomal p... 220 3e-57
sce:YGL135W RPL1B, SSM2; L1B; K02865 large subunit ribosomal p... 220 3e-57
cpv:cgd7_3490 hypothetical protein ; K14775 ribosome biogenesi... 60.1 7e-09
ath:AT1G06380 ribosomal protein-related 57.0 7e-08
ath:AT2G42650 60S ribosomal protein-related 54.7 3e-07
ath:AT3G58660 60S ribosomal protein-related; K14775 ribosome b... 54.7 3e-07
ath:AT2G42710 ribosomal protein L1 family protein 52.0 2e-06
ath:AT3G63490 ribosomal protein L1 family protein 40.4 0.006
cel:F53F4.11 hypothetical protein 40.0 0.007
bbo:BBOV_IV009670 23.m06191; hypothetical protein; K14775 ribo... 37.0 0.061
dre:563247 similar to ribosomal L1 domain containing 1; K14775... 37.0 0.070
xla:100137646 rsl1d1; ribosomal L1 domain containing 1; K14775... 35.0 0.25
ath:AT2G27535 ribosomal protein L10A family protein 31.6 2.6
cel:W04A8.6 hypothetical protein 31.2 3.9
> tgo:TGME49_015470 60S ribosomal protein L10a, putative ; K02865
large subunit ribosomal protein L10Ae
Length=217
Score = 336 bits (862), Expect = 4e-92, Method: Compositional matrix adjust.
Identities = 175/217 (80%), Positives = 194/217 (89%), Gaps = 0/217 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSKLS + L AI IL+GS+EKKRKFVET+ELQI LKDYDTQRDKRFSGSVRLP+VPR
Sbjct 1 MSKLSTDGLKKAIGEILEGSREKKRKFVETVELQIGLKDYDTQRDKRFSGSVRLPNVPRP 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
+ +VCV+GDAVH E+AK LGL+ MDVEA+KK+NKNKK+VKKLARKYDAFLASQ LIPQIP
Sbjct 61 RMRVCVMGDAVHCEQAKELGLEFMDVEAMKKLNKNKKLVKKLARKYDAFLASQVLIPQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
R LGPGLNKAGKFPTLITH DK+EDKI E++SS+KFQLKKVLCMGVAV NVEMTE Q+R
Sbjct 121 RLLGPGLNKAGKFPTLITHNDKLEDKIQEIKSSIKFQLKKVLCMGVAVGNVEMTEEQLRV 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIYG 251
NLT+AINFLVSLLKKNWNNVK LHIKSTMGK IYG
Sbjct 181 NLTLAINFLVSLLKKNWNNVKTLHIKSTMGKPQQIYG 217
> bbo:BBOV_III003180 17.m07305; ribosomal protein L1; K02865 large
subunit ribosomal protein L10Ae
Length=217
Score = 323 bits (828), Expect = 4e-88, Method: Compositional matrix adjust.
Identities = 153/217 (70%), Positives = 184/217 (84%), Gaps = 0/217 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSKL+ ETLN AI++IL SQEKKR FVET+ELQISLKDYDTQRDKRFSG+V L +VPR
Sbjct 1 MSKLTVETLNEAISAILTQSQEKKRNFVETVELQISLKDYDTQRDKRFSGTVVLQNVPRE 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
+ K+CV GD VH ++AK+LG+D +D+E LKK N+NK +VKKLA KY AFLASQ L+PQIP
Sbjct 61 RMKICVFGDQVHCDQAKSLGIDYIDLEGLKKFNRNKTLVKKLANKYGAFLASQTLLPQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
R+LGPGLNKAGKFPT +TH D +E+K+ E++SSVKFQLKKVLCMGVAV NVEMT Q+R
Sbjct 121 RFLGPGLNKAGKFPTQLTHNDNMEEKVREIKSSVKFQLKKVLCMGVAVGNVEMTHEQLRA 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIYG 251
N+ +AIN+LVSLLKKNW+NV+ L +KSTMGK IYG
Sbjct 181 NIVLAINYLVSLLKKNWHNVRGLTVKSTMGKPFRIYG 217
> tpv:TP04_0125 60S ribosomal protein L1; K02865 large subunit
ribosomal protein L10Ae
Length=217
Score = 318 bits (816), Expect = 1e-86, Method: Compositional matrix adjust.
Identities = 151/217 (69%), Positives = 182/217 (83%), Gaps = 0/217 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSKLS + LN A+ ++L+GS+ KKR F ET+ELQISLKDYDTQRDKRFSG+V L + P+
Sbjct 1 MSKLSSDRLNDAVTAVLEGSKTKKRNFTETVELQISLKDYDTQRDKRFSGTVVLQNAPKK 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
KVCV GDAVH +EAKALG+D +D+E LKK N+NK +VKKLA KY AFLASQ+L+PQIP
Sbjct 61 NMKVCVFGDAVHCDEAKALGVDFIDLEGLKKFNRNKTLVKKLANKYSAFLASQSLLPQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
R+LGPGLNKAGKFPT + H D++EDKI E+RSSVKFQLKKVLCMGVAV NVEM+ Q++
Sbjct 121 RFLGPGLNKAGKFPTQLLHTDRMEDKINELRSSVKFQLKKVLCMGVAVGNVEMSPEQLKA 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIYG 251
N+ +++N+LVSLLKKNWNNVK L IKSTMGK IYG
Sbjct 181 NIVLSVNYLVSLLKKNWNNVKGLTIKSTMGKPQRIYG 217
> pfa:PF14_0391 60S ribosomal protein L1, putative; K02865 large
subunit ribosomal protein L10Ae
Length=217
Score = 318 bits (815), Expect = 1e-86, Method: Compositional matrix adjust.
Identities = 150/217 (69%), Positives = 184/217 (84%), Gaps = 0/217 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSKL+Q+ L AI+ + +G+++KKRKFVETIELQI LKDYDTQRDKRFSG+VRL + R
Sbjct 1 MSKLNQDLLKKAISDVFEGTKQKKRKFVETIELQIGLKDYDTQRDKRFSGTVRLSNEVRK 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
K KVC++GDAVH+EEA+ L LD MD+EA+KK+NK+K +VKKLA+KYDAFLASQ ++PQIP
Sbjct 61 KLKVCILGDAVHVEEAQKLELDYMDIEAMKKLNKDKTLVKKLAKKYDAFLASQVILPQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
+ LGPGLNKAGKFP+LITH DKI DKILE++SS+KFQLKKVLCMGV V + + E ++R
Sbjct 121 KLLGPGLNKAGKFPSLITHNDKINDKILELKSSIKFQLKKVLCMGVPVGHANLKEEELRS 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIYG 251
N+ AINFLVSLLKKNW N++ LHIKSTMGK IYG
Sbjct 181 NIVHAINFLVSLLKKNWQNIRTLHIKSTMGKPQRIYG 217
> cpv:cgd8_2870 60S ribosomal protein L10A ; K02865 large subunit
ribosomal protein L10Ae
Length=221
Score = 285 bits (728), Expect = 1e-76, Method: Compositional matrix adjust.
Identities = 148/216 (68%), Positives = 181/216 (83%), Gaps = 0/216 (0%)
Query 36 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 95
K+S ET+ AI+ IL+GS+ K RKFVET+ELQI LKDYDTQRDKRF+G+VRLP+VPR
Sbjct 6 GKVSSETIRRAISEILEGSKAKPRKFVETVELQIGLKDYDTQRDKRFAGTVRLPNVPRPN 65
Query 96 SKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIPR 155
++VCV+GDA E A+ LG D MD+E +KKINKNKK+VKKL +KYD FLASQ L+PQIPR
Sbjct 66 ARVCVMGDAADCERAQKLGFDVMDIEEMKKINKNKKVVKKLCKKYDLFLASQVLLPQIPR 125
Query 156 YLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRYN 215
LGPGLNKAGKFPT+IT DKI++K E+++S+KFQLKKVLC+GVA+ NV MTE ++R N
Sbjct 126 LLGPGLNKAGKFPTVITPSDKIDEKANELKASIKFQLKKVLCLGVAIGNVNMTEEEIRQN 185
Query 216 LTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIYG 251
LT+AINFLVSLLKKNW+N+K L +KSTMGK IYG
Sbjct 186 LTLAINFLVSLLKKNWHNIKSLTVKSTMGKSVRIYG 221
> ath:AT1G08360 60S ribosomal protein L10A (RPL10aA); K02865 large
subunit ribosomal protein L10Ae
Length=216
Score = 266 bits (679), Expect = 7e-71, Method: Compositional matrix adjust.
Identities = 140/216 (64%), Positives = 178/216 (82%), Gaps = 0/216 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSKL E + AI +I S+ KKR FVETIELQI LK+YD Q+DKRFSGSV+LPH+PR
Sbjct 1 MSKLQSEAVREAITTITGKSEAKKRNFVETIELQIGLKNYDPQKDKRFSGSVKLPHIPRP 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
K K+C++GDA H+EEA+ +GL+ MDVE+LKK+NKNKK+VKKLA+KY AFLAS+++I QIP
Sbjct 61 KMKICMLGDAQHVEEAEKMGLENMDVESLKKLNKNKKLVKKLAKKYHAFLASESVIKQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
R LGPGLNKAGKFPTL++HQ+ +E K+ E +++VKFQLKKVLCMGVAV N+ M E Q+
Sbjct 121 RLLGPGLNKAGKFPTLVSHQESLESKVNETKATVKFQLKKVLCMGVAVGNLSMEEKQIFQ 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
N+ M++NFLVSLLKKNW NV+ L++KSTMG I+
Sbjct 181 NVQMSVNFLVSLLKKNWQNVRCLYLKSTMGPPQRIF 216
> ath:AT2G27530 PGY1; PGY1 (PIGGYBACK1); RNA binding / structural
constituent of ribosome
Length=216
Score = 264 bits (674), Expect = 2e-70, Method: Compositional matrix adjust.
Identities = 140/216 (64%), Positives = 177/216 (81%), Gaps = 0/216 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSKL E + AI +I S+EKKR FVET+ELQI LK+YD Q+DKRFSGSV+LPH+PR
Sbjct 1 MSKLQSEAVREAITTIKGKSEEKKRNFVETVELQIGLKNYDPQKDKRFSGSVKLPHIPRP 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
K K+C++GDA H+EEA+ +GL MDVEALKK+NKNKK+VKKLA+ Y AFLAS+++I QIP
Sbjct 61 KMKICMLGDAQHVEEAEKMGLSNMDVEALKKLNKNKKLVKKLAKSYHAFLASESVIKQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
R LGPGLNKAGKFPTL++HQ+ +E K+ E +++VKFQLKKVLCMGVAV N+ M E Q+
Sbjct 121 RLLGPGLNKAGKFPTLVSHQESLEAKVNETKATVKFQLKKVLCMGVAVGNLSMEEKQLFQ 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
N+ M++NFLVSLLKKNW NV+ L++KSTMG I+
Sbjct 181 NVQMSVNFLVSLLKKNWQNVRCLYLKSTMGPPQRIF 216
> ath:AT5G22440 60S ribosomal protein L10A (RPL10aC); K02865 large
subunit ribosomal protein L10Ae
Length=217
Score = 259 bits (661), Expect = 9e-69, Method: Compositional matrix adjust.
Identities = 139/211 (65%), Positives = 176/211 (83%), Gaps = 1/211 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKK-RKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPR 93
MSKL E + AI+SI+ +E K R F ETIELQI LK+YD Q+DKRFSGSV+LPHVPR
Sbjct 1 MSKLQSEAVREAISSIITHCKETKPRNFTETIELQIGLKNYDPQKDKRFSGSVKLPHVPR 60
Query 94 AKSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQI 153
K K+C++GDA H+EEA+ +GL+ MDVEALKK+NKNKK+VKKLA+K+ AFLAS+++I QI
Sbjct 61 PKMKICMLGDAQHVEEAEKIGLESMDVEALKKLNKNKKLVKKLAKKFHAFLASESVIKQI 120
Query 154 PRYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVR 213
PR LGPGLNKAGKFPTL++HQ+ +E K+ E +++VKFQLKKVLCMGVAV N+ M E Q+
Sbjct 121 PRLLGPGLNKAGKFPTLVSHQESLESKVNETKATVKFQLKKVLCMGVAVGNLSMEEKQIF 180
Query 214 YNLTMAINFLVSLLKKNWNNVKRLHIKSTMG 244
N+ M++NFLVSLLKKNW NV+ L++KSTMG
Sbjct 181 QNVQMSVNFLVSLLKKNWQNVRCLYLKSTMG 211
> xla:379897 rpl10a, MGC53614; ribosomal protein L10a (EC:3.6.5.3);
K02865 large subunit ribosomal protein L10Ae
Length=217
Score = 254 bits (650), Expect = 2e-67, Method: Compositional matrix adjust.
Identities = 137/215 (63%), Positives = 181/215 (84%), Gaps = 0/215 (0%)
Query 36 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 95
SK+S++TL A+ +L+GS++KKRKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR K
Sbjct 3 SKVSRDTLYEAVREVLQGSKKKKRKFLETVELQISLKNYDPQKDKRFSGTVRLKSTPRPK 62
Query 96 SKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIPR 155
+CV+GD H +EAKA+ L MD+EALKK+NKNKK+VKKLA+KYDAFLAS++LI QIPR
Sbjct 63 FSLCVLGDQQHCDEAKAVDLPHMDIEALKKLNKNKKLVKKLAKKYDAFLASESLIKQIPR 122
Query 156 YLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRYN 215
LGPGLNKAGKFP+L+TH + + K+ EV+S++KFQ+KKVLC+ VAV +V++TE ++ YN
Sbjct 123 ILGPGLNKAGKFPSLLTHNENLVAKVDEVKSTIKFQMKKVLCLAVAVGHVKLTEEELVYN 182
Query 216 LTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
+ ++INFLVSLLKKNW NV+ L+IKSTMGK +Y
Sbjct 183 IHLSINFLVSLLKKNWQNVRALYIKSTMGKPQRLY 217
> dre:323305 rpl10a, MGC86881, wu:fb94f08, zgc:73082, zgc:86881;
ribosomal protein L10a; K02865 large subunit ribosomal protein
L10Ae
Length=216
Score = 254 bits (649), Expect = 2e-67, Method: Compositional matrix adjust.
Identities = 138/216 (63%), Positives = 178/216 (82%), Gaps = 0/216 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSK+S++TL A+ + GS+ KKRKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR
Sbjct 1 MSKVSRDTLYEAVKEVQAGSRRKKRKFLETVELQISLKNYDPQKDKRFSGTVRLKTTPRP 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
K VCV+GD H +EAKA L MD+EALKK+NKNKK+VKKLA+KYDAFLAS++LI QIP
Sbjct 61 KFSVCVLGDQQHCDEAKAAELPHMDIEALKKLNKNKKLVKKLAKKYDAFLASESLIKQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
R LGPGLNKAGKFP+L+TH + + K+ EV+S++KFQ+KKVLC+ VAV +V+M+E ++ Y
Sbjct 121 RILGPGLNKAGKFPSLLTHNENLGTKVDEVKSTIKFQMKKVLCLAVAVGHVKMSEEELVY 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
N+ +A+NFLVSLLKKNW NV+ L+IKSTMGK +Y
Sbjct 181 NIHLAVNFLVSLLKKNWQNVRALYIKSTMGKPQRLY 216
> mmu:19896 Rpl10a, CsA-19, Nedd6; ribosomal protein L10A (EC:3.6.5.3);
K02865 large subunit ribosomal protein L10Ae
Length=217
Score = 253 bits (647), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 136/215 (63%), Positives = 179/215 (83%), Gaps = 0/215 (0%)
Query 36 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 95
SK+S++TL A+ +L G+Q K+RKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR K
Sbjct 3 SKVSRDTLYEAVREVLHGNQRKRRKFLETVELQISLKNYDPQKDKRFSGTVRLKSTPRPK 62
Query 96 SKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIPR 155
VCV+GD H +EAKA+ + MD+EALKK+NKNKK+VKKLA+KYDAFLAS++LI QIPR
Sbjct 63 FSVCVLGDQQHCDEAKAVDIPHMDIEALKKLNKNKKLVKKLAKKYDAFLASESLIKQIPR 122
Query 156 YLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRYN 215
LGPGLNKAGKFP+L+TH + + K+ EV+S++KFQ+KKVLC+ VAV +V+MT+ ++ YN
Sbjct 123 ILGPGLNKAGKFPSLLTHNENMVAKVDEVKSTIKFQMKKVLCLAVAVGHVKMTDDELVYN 182
Query 216 LTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
+ +A+NFLVSLLKKNW NV+ L+IKSTMGK +Y
Sbjct 183 IHLAVNFLVSLLKKNWQNVRALYIKSTMGKPQRLY 217
> hsa:4736 RPL10A, Csa-19, NEDD6; ribosomal protein L10a; K02865
large subunit ribosomal protein L10Ae
Length=217
Score = 253 bits (647), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 136/215 (63%), Positives = 179/215 (83%), Gaps = 0/215 (0%)
Query 36 SKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAK 95
SK+S++TL A+ +L G+Q K+RKF+ET+ELQISLK+YD Q+DKRFSG+VRL PR K
Sbjct 3 SKVSRDTLYEAVREVLHGNQRKRRKFLETVELQISLKNYDPQKDKRFSGTVRLKSTPRPK 62
Query 96 SKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIPR 155
VCV+GD H +EAKA+ + MD+EALKK+NKNKK+VKKLA+KYDAFLAS++LI QIPR
Sbjct 63 FSVCVLGDQQHCDEAKAVDIPHMDIEALKKLNKNKKLVKKLAKKYDAFLASESLIKQIPR 122
Query 156 YLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRYN 215
LGPGLNKAGKFP+L+TH + + K+ EV+S++KFQ+KKVLC+ VAV +V+MT+ ++ YN
Sbjct 123 ILGPGLNKAGKFPSLLTHNENMVAKVDEVKSTIKFQMKKVLCLAVAVGHVKMTDDELVYN 182
Query 216 LTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
+ +A+NFLVSLLKKNW NV+ L+IKSTMGK +Y
Sbjct 183 IHLAVNFLVSLLKKNWQNVRALYIKSTMGKPQRLY 217
> cel:Y71F9AL.13 rpl-1; Ribosomal Protein, Large subunit family
member (rpl-1); K02865 large subunit ribosomal protein L10Ae
Length=216
Score = 253 bits (647), Expect = 4e-67, Method: Compositional matrix adjust.
Identities = 132/216 (61%), Positives = 174/216 (80%), Gaps = 0/216 (0%)
Query 35 MSKLSQETLNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRA 94
MSK+S+E+LN AIA +LKGS EK RKF ETIELQI LK+YD Q+DKRFSGS+RL H+PR
Sbjct 1 MSKVSRESLNEAIAEVLKGSSEKPRKFRETIELQIGLKNYDPQKDKRFSGSIRLKHIPRP 60
Query 95 KSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP 154
KVCV GD H++EA A + M + LKK+NK KK++KKLA+ YDAF+AS++LI QIP
Sbjct 61 NMKVCVFGDQHHLDEAAAGDIPSMSADDLKKLNKQKKLIKKLAKSYDAFIASESLIKQIP 120
Query 155 RYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRY 214
R LGPGLNKAGKFP+++TH + ++ K E+R++VKFQ+KKVLC+ VAV +V +T+ ++
Sbjct 121 RILGPGLNKAGKFPSVVTHGESLQSKSDEIRATVKFQMKKVLCLSVAVGHVGLTQEELVS 180
Query 215 NLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
N++++INFLVSLLKKNW NV+ L+IKSTMGK +Y
Sbjct 181 NISLSINFLVSLLKKNWQNVRSLNIKSTMGKPQRVY 216
> sce:YPL220W RPL1A, SSM1; L1A; K02865 large subunit ribosomal
protein L10Ae
Length=217
Score = 220 bits (561), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 118/217 (54%), Positives = 162/217 (74%), Gaps = 1/217 (0%)
Query 35 MSKLSQETLNAAIASILKGSQE-KKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPR 93
MSK++ + + +LK S E KKR F+ET+ELQ+ LK+YD QRDKRFSGS++LP+ PR
Sbjct 1 MSKITSSQVREHVKELLKYSNETKKRNFLETVELQVGLKNYDPQRDKRFSGSLKLPNCPR 60
Query 94 AKSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQI 153
+C+ GDA ++ AK+ G+D M V+ LKK+NKNKK++KKL++KY+AF+AS+ LI Q+
Sbjct 61 PNMSICIFGDAFDVDRAKSCGVDAMSVDDLKKLNKNKKLIKKLSKKYNAFIASEVLIKQV 120
Query 154 PRYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVR 213
PR LGP L+KAGKFPT ++H D + K+ +VRS++KFQLKKVLC+ VAV NVEM E +
Sbjct 121 PRLLGPQLSKAGKFPTPVSHNDDLYGKVTDVRSTIKFQLKKVLCLAVAVGNVEMEEDVLV 180
Query 214 YNLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
+ M++NF VSLLKKNW NV L +KS+MG +Y
Sbjct 181 NQILMSVNFFVSLLKKNWQNVGSLVVKSSMGPAFRLY 217
> sce:YGL135W RPL1B, SSM2; L1B; K02865 large subunit ribosomal
protein L10Ae
Length=217
Score = 220 bits (561), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 118/217 (54%), Positives = 162/217 (74%), Gaps = 1/217 (0%)
Query 35 MSKLSQETLNAAIASILKGSQE-KKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPR 93
MSK++ + + +LK S E KKR F+ET+ELQ+ LK+YD QRDKRFSGS++LP+ PR
Sbjct 1 MSKITSSQVREHVKELLKYSNETKKRNFLETVELQVGLKNYDPQRDKRFSGSLKLPNCPR 60
Query 94 AKSKVCVIGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQI 153
+C+ GDA ++ AK+ G+D M V+ LKK+NKNKK++KKL++KY+AF+AS+ LI Q+
Sbjct 61 PNMSICIFGDAFDVDRAKSCGVDAMSVDDLKKLNKNKKLIKKLSKKYNAFIASEVLIKQV 120
Query 154 PRYLGPGLNKAGKFPTLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVR 213
PR LGP L+KAGKFPT ++H D + K+ +VRS++KFQLKKVLC+ VAV NVEM E +
Sbjct 121 PRLLGPQLSKAGKFPTPVSHNDDLYGKVTDVRSTIKFQLKKVLCLAVAVGNVEMEEDVLV 180
Query 214 YNLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
+ M++NF VSLLKKNW NV L +KS+MG +Y
Sbjct 181 NQILMSVNFFVSLLKKNWQNVGSLVVKSSMGPAFRLY 217
> cpv:cgd7_3490 hypothetical protein ; K14775 ribosome biogenesis
protein UTP30
Length=250
Score = 60.1 bits (144), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 60/115 (52%), Gaps = 6/115 (5%)
Query 134 KKLARKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLI-THQDKIEDKILEVRSSVKFQL 192
++L YD FL+ +I ++P+ LG KA K P I + ++ + SS +
Sbjct 124 RELCSGYDLFLSDDRIIEKMPKLLGSYFIKANKMPIAIKIRESNFKNSLESALSSTFMSI 183
Query 193 KKVLCMGVAVANVEMTEAQVRYNLTMAI----NFLVSLLKKNW-NNVKRLHIKST 242
KK C+G+ VA V+M QV NL AI +F KNW N+++ L+I+ST
Sbjct 184 KKGKCIGIRVARVDMNTKQVVQNLIEAIKQVSDFFEGNGSKNWRNSIESLYIQST 238
> ath:AT1G06380 ribosomal protein-related
Length=254
Score = 57.0 bits (136), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 61/232 (26%), Positives = 108/232 (46%), Gaps = 22/232 (9%)
Query 30 FPTPKMSKLSQETLNAAIASILKGSQEK-KRKFVETIE------LQISLKDYDTQRDKRF 82
P SK+ + +N A+ S+LK K K + E++E L ++LK Q D+
Sbjct 10 LPQRHQSKVDPQNVNRAVKSLLKWWDSKSKTENSESLENDGFVYLIVTLKRI-PQLDRTN 68
Query 83 SGSVRLPH-----VPRAKSKVCVIGDAVH----MEEA--KALGLDCMDVEALKKINKNKK 131
+ LPH V ++C+I D H +EA K + + + + + K++K K
Sbjct 69 PLMIPLPHPLIDLVAEDPPELCLIIDDKHKNKITKEAALKKIEAEKIPITTVIKVSKLKS 128
Query 132 IVKKLA--RKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLIT-HQDKIEDKILEVRSSV 188
++KL ++++ + A + L+P +P+ LG K K P I +++I + S
Sbjct 129 DLRKLEEEKRFELYFAERRLMPMLPKLLGKEFVKKNKTPIAINLRHGSWKEQIEKACESA 188
Query 189 KFQLKKVLCMGVAVANVEMTEAQVRYNLTMAINFLVSLLKKNWNNVKRLHIK 240
F + C V VA + M ++ N+ A+N + L+ W NVK H+K
Sbjct 189 LFFVGTGTCSVVKVAKLSMGRNEIAENVVAAMNGIGDLVPGRWKNVKLFHLK 240
> ath:AT2G42650 60S ribosomal protein-related
Length=372
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 55/231 (23%), Positives = 110/231 (47%), Gaps = 27/231 (11%)
Query 35 MSKLSQETLNAAIASILKGSQEKKR----KFVET---IELQISLKDYDTQRDKRFSGSVR 87
MS++S +T++ A+ +++K EK R + +E L ++LK QR+ + +
Sbjct 1 MSRVSPKTVDDAVKALVKEGNEKSRTEKPQLLEEDGFFYLVVALKKI-PQRNFTNAYRIP 59
Query 88 LPH----VPRAKSKVCVIGD-----AVHMEEAKA-LGLDCMDVEALKKINKNKKIV---- 133
LPH ++C+I D + E+AK + + + + + K++K K
Sbjct 60 LPHPLINTTEDSPELCLIIDDRPESGLTEEDAKKNIKSENIPITKVVKLSKLKSDYGSFE 119
Query 134 --KKLARKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLIT--HQDKIEDKILEVRSSVK 189
+KL YD F + + +IP +P+ +G ++ K P I H + +++I + +
Sbjct 120 SKRKLCDSYDMFFSDRRVIPMLPKLIGKKFFQSKKTPVAIDLKHMN-WKEQIEKACGAAM 178
Query 190 FQLKKVLCMGVAVANVEMTEAQVRYNLTMAINFLVSLLKKNWNNVKRLHIK 240
F ++ C + VA + M + N+T +N +V +L W ++ LH+K
Sbjct 179 FFMRTGSCSAIKVAKLSMESDDIVENVTATLNGVVDVLPSRWKYIRSLHLK 229
> ath:AT3G58660 60S ribosomal protein-related; K14775 ribosome
biogenesis protein UTP30
Length=446
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 60/254 (23%), Positives = 113/254 (44%), Gaps = 26/254 (10%)
Query 22 PPDPLERRF-PTPKMSKLSQETLNAAIASILKG----SQEKKRKFVETIELQ---ISLKD 73
PP P E++ + S++S +T+ +A+ ++ S+ +K + +E EL ++LK
Sbjct 7 PPPPQEQQLVHASQTSRVSPKTVESALNGLINWRSDKSKTEKPQLLEEDELVYLFVTLKK 66
Query 74 YDTQRDKRFSGSVRLPHV---PRAKS-KVCVIGDA---------VHMEEAKALGLDCMDV 120
Q+ + + + LPH P S ++C+I D M++ K+ + V
Sbjct 67 I-PQKTRTNAYRIPLPHPLINPTVDSPEICLIIDDRPKSGLTKDDAMKKIKSENIPITKV 125
Query 121 EALKKINKNKKIV---KKLARKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLITHQDK- 176
L K+ + K +KL YD F + +IP +PR +G + K P + + +
Sbjct 126 IKLSKLKSDYKAFEAKRKLCDSYDMFFTDRRIIPLLPRVIGKKFFTSKKIPVALDLKHRN 185
Query 177 IEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRYNLTMAINFLVSLLKKNWNNVKR 236
+ +I + S F ++ C + V + M ++ N+ +N LV L W V+
Sbjct 186 WKHQIEKACGSAMFFIRTGTCSVIKVGKLSMDICEITENVMATLNGLVEFLPNKWTYVRS 245
Query 237 LHIKSTMGKCHTIY 250
LH+K + IY
Sbjct 246 LHLKLSESLALPIY 259
> ath:AT2G42710 ribosomal protein L1 family protein
Length=415
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 52/205 (25%), Positives = 92/205 (44%), Gaps = 22/205 (10%)
Query 58 KRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAKSKVCVIGDAVHMEEAKALGLDC 117
K KF ET+E + L + + G++ LPH + KV + E+AKA G D
Sbjct 202 KAKFDETLEAHVRLGIEKGRSELIVRGTLALPHSVKKDVKVAFFAEGADAEDAKAAGADV 261
Query 118 M-DVEALKKINKNKKIVKKLARKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLITHQDK 176
+ +E +++I K+ KI +D LA+ ++P++ + + LN G P Q
Sbjct 262 VGGLELIEEILKSGKI------DFDRCLATPKMMPRVYK-ISRILNNHGLMPN--PKQGS 312
Query 177 IEDKILEV-----RSSVKFQLKKVLCMGVAVANVEMTEAQVRYNLTMAINFLV----SLL 227
+ + + KF++ K + V + + +E +R N+ +N L+ + L
Sbjct 313 VTKDVTKAVKDAKAGHTKFRMDKTSILHVPLGKMSFSEEALRENVGAFMNALLLAKPAGL 372
Query 228 KKN---WNNVKRLHIKSTMGKCHTI 249
KK V H+ STMGK + +
Sbjct 373 KKTSKYAGYVNAFHLCSTMGKGYPV 397
> ath:AT3G63490 ribosomal protein L1 family protein
Length=346
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 54/209 (25%), Positives = 90/209 (43%), Gaps = 19/209 (9%)
Query 43 LNAAIASILKGSQEKKRKFVETIELQISLKDYDTQRDKRFSGSVRLPHVPRAKSKVCVIG 102
+N AI S+LK Q +FVE++E L D++ +V LP V V+
Sbjct 131 VNTAI-SLLK--QTANTRFVESVEAHFRLNIDPKYNDQQLRATVSLPKGTGQTVIVAVLA 187
Query 103 DAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIP---RYLGP 159
++EAK+ G D + + L + +K ++D +AS ++ ++ + LGP
Sbjct 188 QGEKVDEAKSAGADIVGSDDL------IEQIKGGFMEFDKLIASPDMMVKVAGLGKILGP 241
Query 160 -GLNKAGKFPTLITHQDKIEDKILEV-RSSVKFQLKKVLCMGVAVANVEMTEAQVRYNLT 217
GL K T+ + I I E + V+F+ K + + V TE + N
Sbjct 242 RGLMPNPKAGTVTAN---IPQAIEEFKKGKVEFRADKTGIVHIPFGKVNFTEEDLLINFL 298
Query 218 MAINFLVSLLKKNWNNV--KRLHIKSTMG 244
A+ + + K V K HI S+MG
Sbjct 299 AAVKSVETNKPKGAKGVYWKSAHICSSMG 327
> cel:F53F4.11 hypothetical protein
Length=543
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 28/128 (21%), Positives = 59/128 (46%), Gaps = 4/128 (3%)
Query 119 DVEALKKINKNKKIVKKLARKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLITHQDKIE 178
+VE + K+K+ LA YD FL+ + + +LG +A K P +Q I
Sbjct 286 EVERIAHTYKDKR---SLASTYDVFLSDGRVYNSVKSFLGKEFYRAHKCPLPFVYQKPIS 342
Query 179 DKILEVRSSVKFQLKKVLCMG-VAVANVEMTEAQVRYNLTMAINFLVSLLKKNWNNVKRL 237
I +V + L++ + V V ++ + A ++ N+ + + S + N++ +
Sbjct 343 TAIENALRTVVYPLRRYMVRSCVNVGHLGQSSADLKENIDTVLEKIASKCPGGFANIRSI 402
Query 238 HIKSTMGK 245
++ ++ GK
Sbjct 403 YLGASDGK 410
> bbo:BBOV_IV009670 23.m06191; hypothetical protein; K14775 ribosome
biogenesis protein UTP30
Length=311
Score = 37.0 bits (84), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 54/122 (44%), Gaps = 6/122 (4%)
Query 101 IGDAVHMEEAKALGLDCMDVEALKKINKNKKIVKKLARKYDAFLASQALIPQIPRYLGPG 160
+GD E K +G VE L+K K K + L +D FL+ Q + P +P LG
Sbjct 139 LGDLKIREIQKVIG-----VEKLRKKYKEYKDRRLLVNSFDLFLSDQRVAPSLPTLLGKI 193
Query 161 LNKAGKFP-TLITHQDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRYNLTMA 219
+ K P +L + + D I+ +S +++ C V VA M+ +V N+
Sbjct 194 FIEKKKMPISLTLGRGSMRDHIVSAINSTFYRVGIGKCSSVKVAVSSMSVDEVVANVQDC 253
Query 220 IN 221
I
Sbjct 254 IE 255
> dre:563247 similar to ribosomal L1 domain containing 1; K14775
ribosome biogenesis protein UTP30
Length=373
Score = 37.0 bits (84), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 55/120 (45%), Gaps = 4/120 (3%)
Query 134 KKLARKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLITHQDKIEDKILE---VRSSVKF 190
+KL +D FL+ + ++P +G + K P + + K + +E +++
Sbjct 136 RKLLGNFDLFLSDARIRFRLPSQIGKHFYERKKAPLSVDLESKHLARDMERLIQGTTLTV 195
Query 191 QLKKVLCMGVAVANVEMTEAQVRYNLTMAINFLVSLLKKNWNNVKRLHIKSTMGKCHTIY 250
K CM VA+ EMT Q+ N+ A++ + + L N+K +H+KS IY
Sbjct 196 STKGPCCMA-RVAHSEMTADQIVENVISAVSTISAKLASTGKNIKIIHLKSQTSVALPIY 254
> xla:100137646 rsl1d1; ribosomal L1 domain containing 1; K14775
ribosome biogenesis protein UTP30
Length=318
Score = 35.0 bits (79), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 53/128 (41%), Gaps = 5/128 (3%)
Query 119 DVEALKKINKNKKIV---KKLARKYDAFLASQALIPQIPRYLGPGLNKAGKFPTLITHQD 175
+V ALK++ K K ++L +D FL+ + +P LG KA + P + +
Sbjct 106 EVIALKRLKKEYKPYEAKRRLLASFDLFLSDARIRRFLPSLLGKHFYKAKREPQSVNLKS 165
Query 176 KIEDKILE--VRSSVKFQLKKVLCMGVAVANVEMTEAQVRYNLTMAINFLVSLLKKNWNN 233
+L V+ + K C + V + +M + N L L W N
Sbjct 166 NHLAAVLNRFVQGTQLHINNKGCCYSIRVGHTDMKVGDIVENAVAVAKVLAEKLPMKWKN 225
Query 234 VKRLHIKS 241
VK LH+K+
Sbjct 226 VKILHLKT 233
> ath:AT2G27535 ribosomal protein L10A family protein
Length=81
Score = 31.6 bits (70), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 30/48 (62%), Gaps = 5/48 (10%)
Query 21 RPPDPLERRFPTPKMSKLSQETLNAAIASILKGSQEKKRKFVETIELQ 68
RP + ER+ KM +L E+L AIA+I S+EKKR +T++LQ
Sbjct 18 RPSE--ERKL---KMRQLESESLREAIATIEGKSEEKKRNVGKTVKLQ 60
> cel:W04A8.6 hypothetical protein
Length=739
Score = 31.2 bits (69), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 174 QDKIEDKILEVRSSVKFQLKKVLCMGVAVANVEMTEAQVRYNLTM-AINFLVSLLKK 229
Q + D + R ++ + + L G+A ANVE+ E +Y LTM A F ++L+K+
Sbjct 134 QINVTDDVDGWRLDMQLRNRHELAPGIAEANVELEEGGKKYRLTMSAGEFKLALVKQ 190
Lambda K H
0.318 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9020732748
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40