bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3563_orf1
Length=411
Score E
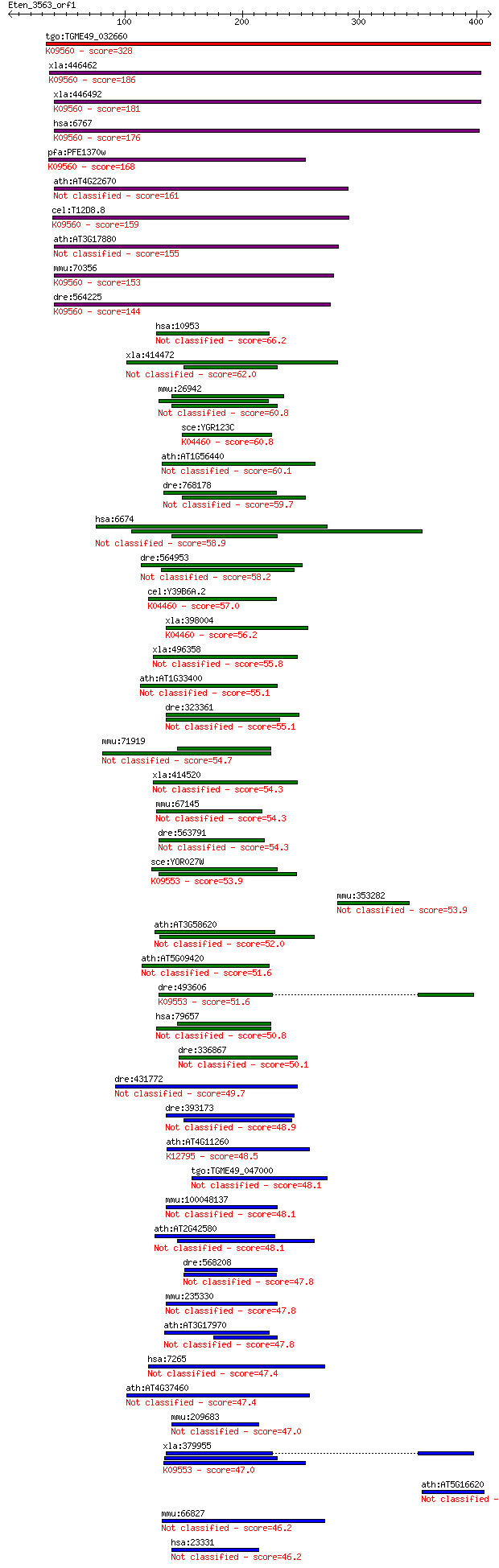
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_032660 58 kDa phosphoprotein, putative ; K09560 sup... 328 2e-89
xla:446462 st13, MGC78939; suppression of tumorigenicity 13 (c... 186 1e-46
xla:446492 MGC79131 protein; K09560 suppressor of tumorigenici... 181 6e-45
hsa:6767 ST13, AAG2, FAM10A1, FAM10A4, FLJ27260, HIP, HOP, HSP... 176 1e-43
pfa:PFE1370w hsp70 interacting protein, putative; K09560 suppr... 168 3e-41
ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-inter... 161 4e-39
cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorig... 159 2e-38
ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING... 155 2e-37
mmu:70356 St13, 1110007I03Rik, 3110002K08Rik, AW555194, HIP, H... 153 1e-36
dre:564225 st13, MGC73267, MGC77089, wu:fd15g02, zgc:73267; su... 144 7e-34
hsa:10953 TOMM34, HTOM34P, TOM34, URCC3; translocase of outer ... 66.2 2e-10
xla:414472 rpap3, MGC81126; RNA polymerase II associated prote... 62.0 4e-09
mmu:26942 Spag1, tpis; sperm associated antigen 1 60.8
sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphat... 60.8 1e-08
ath:AT1G56440 serine/threonine protein phosphatase-related 60.1 1e-08
dre:768178 zgc:153288 59.7 2e-08
hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associate... 58.9 4e-08
dre:564953 spag1, MGC162178, cb1089, wu:fj78g10; sperm associa... 58.2 7e-08
cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);... 57.0 1e-07
xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subuni... 56.2 2e-07
xla:496358 sgta; small glutamine-rich tetratricopeptide repeat... 55.8 3e-07
ath:AT1G33400 tetratricopeptide repeat (TPR)-containing protein 55.1 4e-07
dre:323361 tomm34, wu:fb96b08, zgc:56645; translocase of outer... 55.1 5e-07
mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase II... 54.7 6e-07
xla:414520 hypothetical protein MGC81394 54.3 8e-07
mmu:67145 Tomm34, 2610100K07Rik, TOM34, Tomm34a, Tomm34b; tran... 54.3 9e-07
dre:563791 ttc12; tetratricopeptide repeat domain 12 54.3
sce:YOR027W STI1; Hsp90 cochaperone, interacts with the Ssa gr... 53.9 1e-06
mmu:353282 Sfmbt2, D2Wsu23e, D330030P06Rik; Scm-like with four... 53.9 1e-06
ath:AT3G58620 TTL4; TTL4 (Tetratricopetide-repeat Thioredoxin-... 52.0 4e-06
ath:AT5G09420 ATTOC64-V (ARABIDOPSIS THALIANA TRANSLOCON AT TH... 51.6 5e-06
dre:493606 stip1, zgc:92133; stress-induced-phosphoprotein 1 (... 51.6 5e-06
hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein 3 50.8
dre:336867 fk20d10, wu:fa05b08, wu:fc52b05, wu:fk20d10, zgc:77... 50.1 2e-05
dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopep... 49.7 2e-05
dre:393173 MGC56178; zgc:56178 48.9 3e-05
ath:AT4G11260 SGT1B; SGT1B; protein binding; K12795 suppressor... 48.5 5e-05
tgo:TGME49_047000 TPR domain-containing protein (EC:3.4.21.72) 48.1 5e-05
mmu:100048137 tetratricopeptide repeat protein 12-like 48.1 6e-05
ath:AT2G42580 TTL3; TTL3 (TETRATRICOPETIDE-REPEAT THIOREDOXIN-... 48.1 7e-05
dre:568208 ttc6; tetratricopeptide repeat domain 6 47.8
mmu:235330 Ttc12, E330017O07Rik; tetratricopeptide repeat doma... 47.8 8e-05
ath:AT3G17970 atToc64-III (Arabidopsis thaliana translocon at ... 47.8 9e-05
hsa:7265 TTC1, FLJ46404, TPR1; tetratricopeptide repeat domain 1 47.4
ath:AT4G37460 SRFR1; SRFR1 (SUPPRESSOR OF RPS4-RLD 1); protein... 47.4 1e-04
mmu:209683 Ttc28, 2310015L07Rik, 6030435N04, AI428795, AI85176... 47.0 1e-04
xla:379955 stip1, MGC53256; stress-induced-phosphoprotein 1; K... 47.0 1e-04
ath:AT5G16620 TIC40; TIC40 46.2 2e-04
mmu:66827 Ttc1, 4833412C19Rik, C79328, TPR1; tetratricopeptide... 46.2 2e-04
hsa:23331 TTC28, KIAA1043; tetratricopeptide repeat domain 28 46.2
> tgo:TGME49_032660 58 kDa phosphoprotein, putative ; K09560 suppressor
of tumorigenicity protein 13
Length=425
Score = 328 bits (842), Expect = 2e-89, Method: Compositional matrix adjust.
Identities = 208/430 (48%), Positives = 272/430 (63%), Gaps = 62/430 (14%)
Query 33 MALFGEEKLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIP----------DPLA 82
MA +K+AEL+AF+ + DPSILH+ ELSFFKEYL SL A IP D
Sbjct 1 MAALSPQKVAELKAFIGMCERDPSILHRPELSFFKEYLQSLKAAIPAERQAGAGSPDRPT 60
Query 83 EEPKEESEEEPFHVE---------SLNDEEVVAPESTPPPALAPAGEKELTDDDYEKLSK 133
P E+ + +E SL D EV+ PE++P P LAP GEKELTDD+ +KL K
Sbjct 61 TSPVPEASSDDSSLESEVEEFDEESLKDSEVIPPETSPLPPLAPEGEKELTDDELDKLGK 120
Query 134 AKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALK 193
KE A+ A EAG+ +A+E +T+ALL+GNPTALLYTRRA+VL K KR A +RDCDEALK
Sbjct 121 LKEEASAACEAGNSERALEKFTEALLIGNPTALLYTRRADVLLKLKRPVACIRDCDEALK 180
Query 194 LNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHER 253
LNPD+ARAY+IRG +NR LG W++AHSD++MGQKIDYDE +WD+QKLV++K+K IEEHER
Sbjct 181 LNPDSARAYKIRGKANRLLGKWREAHSDLDMGQKIDYDEGLWDMQKLVDEKFKKIEEHER 240
Query 254 SVQRRREEKERREREKRIREQRASSQKAYEEQKKRE------------------------ 289
+ R+ EE E++ REK R++RA++Q+AYEEQK+R+
Sbjct 241 KIVRKCEEAEKKRREKEARKRRAAAQRAYEEQKQRDAAGSGGFPGGFPEGFPGFGGGAGF 300
Query 290 ---------ASGVPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGG 340
+ G+PG FP GG M+G G G G G VPGG
Sbjct 301 PGGAGGFGYSGGMPGTFPSSSCGG--SGMTGTPGGCCGGMGGGCGGGMPSNSVPGGA--- 355
Query 341 MGGLGGLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFG 400
G L G++NDP+++++F NPKMMAAFQDI++NP +++KY DPEV A+ L KFG
Sbjct 356 ----GNLFGALNDPELKKLFENPKMMAAFQDIMSNPSSISKYASDPEVMAAMGSLTSKFG 411
Query 401 GGMAGGSGIP 410
GG+ G+G P
Sbjct 412 GGVP-GAGFP 420
> xla:446462 st13, MGC78939; suppression of tumorigenicity 13
(colon carcinoma) (Hsp70 interacting protein); K09560 suppressor
of tumorigenicity protein 13
Length=379
Score = 186 bits (472), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 128/383 (33%), Positives = 211/383 (55%), Gaps = 21/383 (5%)
Query 36 FGEEKLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIPDPLAEEPKEESEEE-PF 94
+ K+ EL+ FV L +++P ILH E FF ++L+S+GA +P P +EP +++E P
Sbjct 1 MDQRKVRELQEFVRLCQSNPDILHCPEFKFFADWLISMGASVPAPTNKEPPTHTKKETPV 60
Query 95 HVESLN---------------DEEVVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAAA 139
E D E V P P E+T++ ++ ++ K A
Sbjct 61 KEERTPSPPPKPESEESDIEIDNEGVVPGDDDEPQEMGDESVEVTEEMMDQANEKKVEAI 120
Query 140 EALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNA 199
AL G+L KA+E +T+A+ + A+LY +RA V K ++ AA+RDCD A+ +NPD+A
Sbjct 121 NALGEGELEKAIELFTEAIKLNPRIAILYAKRASVYVKLQKPNAAIRDCDRAIAINPDSA 180
Query 200 RAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERSVQRRR 259
+ Y+ RG ++R LG+W+ + D+ + K+DYDE+ + K V+ + I EH R +R+R
Sbjct 181 QPYKWRGKAHRLLGHWEDSAHDLAIACKLDYDEDASTLLKEVQPRANKIAEHRRKYERKR 240
Query 260 EEKERREREKRIREQRASSQKAYEEQKKREASGVPGGFPGGMPGGFPGEMSGGYPGGAPG 319
EE+E ER++R+++ + +++A E++ R +G GG PG
Sbjct 241 EEREINERKERLKKAKEENERAQREEEARHQAG-----EQFGGFPGGFPGGMPGMGGMPG 295
Query 320 GMSGSMPGGIPGGVPGGMPGGMGGLGGLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNM 379
GG+PG GGM G+ G+ ++DP+V +P++MAAFQD+ NP N+
Sbjct 296 MGGMPGMGGMPGMGGMPGMGGMAGMPGVSDILSDPEVLAAMQDPEVMAAFQDVAQNPANI 355
Query 380 AKYKDDPEVADAISKLMRKFGGG 402
+KY+++P+V + I+KL KFGG
Sbjct 356 SKYQNNPKVMNLITKLSSKFGGS 378
> xla:446492 MGC79131 protein; K09560 suppressor of tumorigenicity
protein 13
Length=376
Score = 181 bits (458), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 127/383 (33%), Positives = 207/383 (54%), Gaps = 32/383 (8%)
Query 40 KLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIP--------------DPLAEE- 84
K+ EL FV L +++P +LH E F ++++S+GA +P P+ EE
Sbjct 5 KVQELREFVRLCQSNPDVLHCPEFKFLTDWIISMGASVPAASSKESPTSTKTETPVQEER 64
Query 85 -----PKEESEEEPFHVESLNDEEVVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAAA 139
PK ESEE + D E V P P E+T++ ++ ++ K A
Sbjct 65 TPTPPPKPESEESDIEI----DNEGVIPGDDDEPQEMGDESAEVTEEMMDQANEKKVEAI 120
Query 140 EALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNA 199
AL G+L K++E +T+A+ + A+LY +RA V + ++ AA+RDCD A+ +NPD+A
Sbjct 121 NALGEGELQKSIELFTEAIKLNPRIAILYAKRASVYVQLQKPNAAIRDCDRAIAINPDSA 180
Query 200 RAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERSVQRRR 259
+ Y+ RG ++R LG+W+ + D+ + K+DYDE+ + K V+ + I EH R +R+R
Sbjct 181 QPYKWRGKAHRLLGHWEDSAHDLAIACKLDYDEDASAMLKEVQPRANKIAEHRRKHERKR 240
Query 260 EEKERREREKRIREQRASSQKAYEEQKKREASGVPGGFPGGMPGGFPGEMSGGYPGGAPG 319
EEKE ++++R+++ + +++A E++ R +G G G G M G G
Sbjct 241 EEKEINDKKERLKKAKEENERAQREEEARHQAGAQFGGFPGGFPGGMPGMGGMPGMGGMP 300
Query 320 GMSGSMPGGIPGGVPGGMPGGMGGLGGLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNM 379
GM G G GM G+ +L DP+V +P++MAAFQD+ NP NM
Sbjct 301 GMGGMP-----GMGGMPGMAGMPGVSDILS---DPEVLTAMQDPEVMAAFQDVAQNPANM 352
Query 380 AKYKDDPEVADAISKLMRKFGGG 402
+KY+++P+V + I+KL KFGG
Sbjct 353 SKYQNNPKVMNLITKLSSKFGGS 375
> hsa:6767 ST13, AAG2, FAM10A1, FAM10A4, FLJ27260, HIP, HOP, HSPABP,
HSPABP1, MGC129952, P48, PRO0786, SNC6; suppression of
tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein);
K09560 suppressor of tumorigenicity protein 13
Length=369
Score = 176 bits (447), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 126/383 (32%), Positives = 206/383 (53%), Gaps = 41/383 (10%)
Query 40 KLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIPDPLAEEPKEESE--------- 90
K+ EL AFV + + DPS+LH +E+ F +E++ S+G K+P P ++ K E
Sbjct 5 KVNELRAFVKMCKQDPSVLHTEEMRFLREWVESMGGKVP-PATQKAKSEENTKEEKPDSK 63
Query 91 --------EEPFHVES---LNDEEVVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAAA 139
+EP ES ++ E V+ P++ P + E+T++ ++ + K AA
Sbjct 64 KVEEDLKADEPSSEESDLEIDKEGVIEPDTDAPQEMGDEN-AEITEEMMDQANDKKVAAI 122
Query 140 EALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNA 199
EAL G+L KA++ +T A+ + A+LY +RA V K ++ AA+RDCD A+++NPD+A
Sbjct 123 EALNDGELQKAIDLFTDAIKLNPRLAILYAKRASVFVKLQKPNAAIRDCDRAIEINPDSA 182
Query 200 RAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERSVQRRR 259
+ Y+ RG ++R LG+W++A D+ + K+DYDE+ + K V+ + + I EH R +R+R
Sbjct 183 QPYKWRGKAHRLLGHWEEAAHDLALACKLDYDEDASAMLKEVQPRAQKIAEHRRKYERKR 242
Query 260 EEKERREREKRIREQRASSQKAYEEQKKREASGVPGGFPGGMPGGFPGEMSGGYPGGAPG 319
EE+E +ER +R+++ R ++A E++ R SG Y G
Sbjct 243 EEREIKERIERVKKAREEHERAQREEEARRQSG------------------AQYGSFPGG 284
Query 320 GMSGSMPGGIPGGVPGGMPGGMGGLGGLLGSV-NDPDVQQVFSNPKMMAAFQDILTNPGN 378
G G G L + +DP+V +P++M AFQD+ NP N
Sbjct 285 FPGGMPGNFPGGMPGMGGGMPGMAGMPGLNEILSDPEVLAAMQDPEVMVAFQDVAQNPAN 344
Query 379 MAKYKDDPEVADAISKLMRKFGG 401
M+KY+ +P+V + ISKL KFGG
Sbjct 345 MSKYQSNPKVMNLISKLSAKFGG 367
> pfa:PFE1370w hsp70 interacting protein, putative; K09560 suppressor
of tumorigenicity protein 13
Length=458
Score = 168 bits (426), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 95/263 (36%), Positives = 144/263 (54%), Gaps = 55/263 (20%)
Query 35 LFGEEKLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIPDPLAEEPKEESEEEPF 94
+ E+K+ +L+ FVTL + +PSIL K EL FFK+++ S G K+ S+++ F
Sbjct 1 MIDEKKVEDLKQFVTLCQENPSILLKPELGFFKKFIESFGGKV-----------SKDKEF 49
Query 95 HVESLNDE--------------------------------------------EVVAPEST 110
+S +DE + + E+
Sbjct 50 FEKSASDESTEEKSDEEEVEEDENDKDDVEEEEEDDVQEDEDEEEDDDERCKDFMVEETI 109
Query 111 PPPALAPAGEKELTDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTR 170
P LAP +++L+D+ E++S K AAE ++ +A+E Y K + G P+A++YT+
Sbjct 110 ECPPLAPIVDEDLSDEVLEEISNLKIEAAELVQDNKFEEALEKYNKIIAFGKPSAMIYTK 169
Query 171 RAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDY 230
RA VL KR A +RDC EAL LN D+A AY++R + R+LG W+ AH+DIE GQKIDY
Sbjct 170 RASVLLSLKRPKACIRDCTEALNLNIDSANAYKVRAKAYRHLGKWECAHADIEQGQKIDY 229
Query 231 DENIWDIQKLVEQKYKIIEEHER 253
DE++W++QKL+E+KYK I E R
Sbjct 230 DEDLWEMQKLIEEKYKKIYEKRR 252
> ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-interacting
protein 1); binding
Length=441
Score = 161 bits (408), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 99/279 (35%), Positives = 155/279 (55%), Gaps = 31/279 (11%)
Query 40 KLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIPDPLAEEPKEESEEEPFHVESL 99
KL+EL+ F+ ++DPS+L LSFF++YL SLGAKIP + EE K +++ F VE
Sbjct 5 KLSELKVFIDQCKSDPSLLTTPSLSFFRDYLESLGAKIPTGVHEEDK-DTKPRSFVVEES 63
Query 100 NDE-----------------------------EVVAPESTPPPALAPAGEKELTDDDYEK 130
+D+ + V P++ PP + + E+TD++ E
Sbjct 64 DDDMDETEEVKPKVEEEEEEDEIVESDVELEGDTVEPDNDPPQKMGDSS-VEVTDENREA 122
Query 131 LSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDE 190
+AK A EAL G+ ++A+E T+A+ + +A++Y RA V K K+ AA+RD +
Sbjct 123 AQEAKGKAMEALSEGNFDEAIEHLTRAITLNPTSAIMYGNRASVYIKLKKPNAAIRDANA 182
Query 191 ALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEE 250
AL++NPD+A+ Y+ RG + LG W +A D+ + IDYDE I + K VE +EE
Sbjct 183 ALEINPDSAKGYKSRGMARAMLGEWAEAAKDLHLASTIDYDEEISAVLKKVEPNAHKLEE 242
Query 251 HERSVQRRREEKERREREKRIREQRASSQKAYEEQKKRE 289
H R R R+E+E ++ E+ +RA +Q AY++ KK E
Sbjct 243 HRRKYDRLRKEREDKKAERDRLRRRAEAQAAYDKAKKEE 281
> cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorigenicity
protein 13
Length=422
Score = 159 bits (402), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 100/276 (36%), Positives = 161/276 (58%), Gaps = 27/276 (9%)
Query 39 EKLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIPD----------PLAEEPKEE 88
+ +A L+ FV + + +P++LH E FFK+YL+SLGA +P P++EEPK+E
Sbjct 2 DHVALLKQFVGMCQANPAVLHAPEFGFFKDYLVSLGATLPPKPADKPEGKCPMSEEPKKE 61
Query 89 S------------EEEPFHVESLNDEEVVAPESTPPPALAPAGE--KELTDDDYEKLSKA 134
+ + E +++E V+ PE P G+ KE T+D+ EK S+
Sbjct 62 TPAAEATPEPEIPKPEEIPFPKIDNEGVIEPEEA---VALPMGDSAKEATEDEIEKASEE 118
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
+ A EA GD + A+ +T A+ +A+L+ +RA VL K KR AA+ DCD+A+ +
Sbjct 119 RGKAQEAFSNGDFDTALTHFTAAIEANPGSAMLHAKRANVLLKLKRPVAAIADCDKAISI 178
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERS 254
NPD+A+ Y+ RG +NR LG W +A +D+ K+DYDE + K VE I+E+ R+
Sbjct 179 NPDSAQGYKFRGRANRLLGKWVEAKTDLATACKLDYDEAANEWLKEVEPNAHKIQEYNRA 238
Query 255 VQRRREEKERREREKRIREQRASSQKAYEEQKKREA 290
V+R++ + E ER +R+R + +++KA EE+ KR+A
Sbjct 239 VERQKADIELAERRERVRRAQEANKKAAEEEAKRQA 274
> ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING
THIOREDOXIN); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / protein binding
Length=373
Score = 155 bits (393), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 101/256 (39%), Positives = 152/256 (59%), Gaps = 19/256 (7%)
Query 40 KLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIPDPLAEEPKEESEEEPFH---- 95
++AEL FV + +PSILH L FFKEYL SLGA++P + + + + P H
Sbjct 6 QVAELRRFVEQLKLNPSILHDPSLVFFKEYLRSLGAQVP-KIEKTAETKPSFSPKHDDDD 64
Query 96 ---VES---LNDEEVVAPESTPPPALAPAGE--KELTDDDYEKLSKAKEAAAEALEAGDL 147
+ES L++ +VV P++ PP P G+ E+TD++ + K A EA+ G
Sbjct 65 DDIMESDVELDNSDVVEPDNEPPQ---PMGDPTAEVTDENRDDAQSEKSKAMEAISDGRF 121
Query 148 NKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGT 207
++A+E TKA+++ +A+LY RA V K K+ AA+RD + AL+ N D+A+ Y+ RG
Sbjct 122 DEAIEHLTKAVMLNPTSAILYATRASVFLKVKKPNAAIRDANVALQFNSDSAKGYKSRGM 181
Query 208 SNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERSVQRRREEKE--RR 265
+ LG W++A +D+ + K+DYDE I + K VE K IEEH R QR R+EKE R
Sbjct 182 AKAMLGQWEEAAADLHVASKLDYDEEIGTMLKKVEPNAKRIEEHRRKYQRLRKEKELQRA 241
Query 266 EREKRIREQRASSQKA 281
ERE+R ++Q A ++A
Sbjct 242 ERERR-KQQEAQEREA 256
> mmu:70356 St13, 1110007I03Rik, 3110002K08Rik, AW555194, HIP,
HOP, HSPABP, HSPABP1, PRO0786, SNC6, p48; suppression of tumorigenicity
13; K09560 suppressor of tumorigenicity protein
13
Length=371
Score = 153 bits (386), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 91/272 (33%), Positives = 154/272 (56%), Gaps = 35/272 (12%)
Query 40 KLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIP---------DPLAEEPKEESE 90
K++EL AFV + R DPS+LH +E+ F +E++ S+G K+P + EE ++++
Sbjct 5 KVSELRAFVKMCRQDPSVLHTEEMRFLREWVESMGGKVPPATHKAKSEENTKEEKRDKTT 64
Query 91 EEPFHVESLNDEE---------VVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAAAEA 141
EE E L+ EE V+ P++ P + E+T++ ++ ++ K AA EA
Sbjct 65 EENIKTEELSSEESDLEIDNEGVIEPDTDAPQEMGDEN-AEITEEMMDEANEKKGAAIEA 123
Query 142 LEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARA 201
L G+L KA++ +T A+ + A+LY +RA V K ++ AA+RDCD A+++NPD+A+
Sbjct 124 LNDGELQKAIDLFTDAIKLNPRLAILYAKRASVFVKLQKPNAAIRDCDRAIEINPDSAQP 183
Query 202 YRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEH---------- 251
Y+ RG ++R LG+W++A D+ + K+DYDE+ + + V+ + + I EH
Sbjct 184 YKWRGKAHRLLGHWEEAAHDLALACKLDYDEDASAMLREVQPRAQKIAEHRRKYERKREE 243
Query 252 ------ERSVQRRREEKERREREKRIREQRAS 277
V++ REE ER +RE+ R Q S
Sbjct 244 REIKERIERVKKAREEHERAQREEEARRQSGS 275
> dre:564225 st13, MGC73267, MGC77089, wu:fd15g02, zgc:73267;
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting
protein); K09560 suppressor of tumorigenicity protein
13
Length=362
Score = 144 bits (363), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 87/272 (31%), Positives = 144/272 (52%), Gaps = 37/272 (13%)
Query 40 KLAELEAFVTLARNDPSILHKQELSFFKEYLLSLGAKIPDPLAEEPKEESEEEPFH---- 95
K+ EL+AFV L +PS+LH E+SFFK +LL +GA IP P + P
Sbjct 5 KVTELKAFVQLCNENPSVLHLPEMSFFKTWLLGMGASIPSPTMNDSSSCKGSCPCDGGAK 64
Query 96 -----------------VESLNDEEVVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAA 138
+++E V+ P++ P + E+T++ ++ ++ K A
Sbjct 65 KAAPKPEPAPPSESEESEIEIDNEGVIEPDTDDPQEMGDFENLEVTEEMMDQANEKKTEA 124
Query 139 AEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDN 198
+AL GDL KA++ +T+A+ + A+LY +RA V K ++ AA+RDCD A+ +NPD+
Sbjct 125 IDALGDGDLQKALDLFTEAIKLNPKLAILYAKRASVYVKMQKPNAAIRDCDRAISINPDS 184
Query 199 ARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEH------- 251
A+ Y+ RG +++ LG+W+++ D+ M K+DYDE + K V+ K I +H
Sbjct 185 AQPYKWRGKAHKLLGHWEESARDLAMACKLDYDEEASAMLKEVQPKANKIIDHRRKYERK 244
Query 252 ---------ERSVQRRREEKERREREKRIREQ 274
+ V++ REE ER +RE+ R+Q
Sbjct 245 REEREIRARQERVKKAREEHERAQREEEARQQ 276
> hsa:10953 TOMM34, HTOM34P, TOM34, URCC3; translocase of outer
mitochondrial membrane 34
Length=309
Score = 66.2 bits (160), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/96 (36%), Positives = 56/96 (58%), Gaps = 0/96 (0%)
Query 127 DYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVR 186
D EK KE E ++ G+ KA+E Y+++LL N + Y+ RA K+Y AV+
Sbjct 189 DVEKARVLKEEGNELVKKGNHKKAIEKYSESLLCSNLESATYSNRALCYLVLKQYTEAVK 248
Query 187 DCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDI 222
DC EALKL+ N +A+ R +++ L ++K + +DI
Sbjct 249 DCTEALKLDGKNVKAFYRRAQAHKALKDYKSSFADI 284
> xla:414472 rpap3, MGC81126; RNA polymerase II associated protein
3
Length=660
Score = 62.0 bits (149), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 50/186 (26%), Positives = 93/186 (50%), Gaps = 7/186 (3%)
Query 102 EEVVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVG 161
E++ S + + G+++ D EK KE ++G ++A+E YT+ +
Sbjct 103 EDIDKDNSNETSSDSECGDEDAITVDTEKALSEKEKGNNYFKSGKYDEAIECYTRGMDAD 162
Query 162 NPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSD 221
A+L T RA F+ K++A A DC+ A+ LN D A+AY RG + L N + A D
Sbjct 163 PYNAILPTNRASAFFRLKKFAVAESDCNLAIALNRDYAKAYARRGAARLALKNLQGAKED 222
Query 222 IEMGQKIDYD--ENIWDIQKLVEQKYKIIEEHERSVQRR-----REEKERREREKRIREQ 274
E ++D + E +++K+ ++ Y + + ++ E+E+++ E + R+Q
Sbjct 223 YEKVLELDANNFEAKNELRKINQELYSSASDVQENMATEAKITVENEEEKKQIEIQQRKQ 282
Query 275 RASSQK 280
+A QK
Sbjct 283 QAIMQK 288
Score = 53.1 bits (126), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 150 AVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSN 209
A+E Y++ + N ALL RA K ++Y A DC A+ L+ +A+ RGT++
Sbjct 303 AIECYSQGMEADNTNALLPANRAMAYLKIQKYKEAEADCTLAISLDASYCKAFARRGTAS 362
Query 210 RYLGNWKQAHSDIEMGQKID 229
LG K+A D EM K+D
Sbjct 363 IMLGKQKEAKEDFEMVLKLD 382
> mmu:26942 Spag1, tpis; sperm associated antigen 1
Length=901
Score = 60.8 bits (146), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/95 (32%), Positives = 58/95 (61%), Gaps = 1/95 (1%)
Query 140 EALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNA 199
EA +GD +AV YT++L PTA+ Y RA+ K +R+++A+ DC++AL+L+P N
Sbjct 222 EAFYSGDYEEAVMYYTRSL-SALPTAIAYNNRAQAEIKLQRWSSALEDCEKALELDPGNV 280
Query 200 RAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENI 234
+A R T+ ++ ++A D+ +++ D ++
Sbjct 281 KALLRRATTYKHQNKLQEAVDDLRKVLQVEPDNDL 315
Score = 38.5 bits (88), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 44/98 (44%), Gaps = 8/98 (8%)
Query 140 EALEAGDLNKAVESYTKALLVGNPT--------ALLYTRRAEVLFKQKRYAAAVRDCDEA 191
E G +A Y+ A+ PT ++LY+ RA K+ ++DC+ A
Sbjct 439 ELFRGGQFAEAAAQYSVAIAQLEPTGSANADELSILYSNRAACYLKEGNCRDCIQDCNRA 498
Query 192 LKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID 229
L+L+P + + R + L ++ A+ D + +ID
Sbjct 499 LELHPFSVKPLLRRAMAYETLEQYRNAYVDYKTVLQID 536
Score = 38.5 bits (88), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 48/94 (51%), Gaps = 1/94 (1%)
Query 129 EKLSKA-KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRD 187
EK+ +A KE + ++ + A+ Y + L + + +YT RA K ++ A D
Sbjct 603 EKMFQALKEEGNQLVKDKNYKDAISKYNECLKINSKACAIYTNRALCYLKLGQFEEAKLD 662
Query 188 CDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSD 221
C++AL+++ +N +A + + L N +++ D
Sbjct 663 CEQALQIDGENVKASHRLALAQKGLENCRESGVD 696
> sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphatase
5 [EC:3.1.3.16]
Length=513
Score = 60.8 bits (146), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 47/76 (61%), Gaps = 0/76 (0%)
Query 149 KAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTS 208
KA+E YT+A+ + + ++ ++ RA FK + +A+ DCDEA+KL+P N +AY R S
Sbjct 30 KAIEKYTEAIDLDSTQSIYFSNRAFAHFKVDNFQSALNDCDEAIKLDPKNIKAYHRRALS 89
Query 209 NRYLGNWKQAHSDIEM 224
L +K+A D+ +
Sbjct 90 CMALLEFKKARKDLNV 105
> ath:AT1G56440 serine/threonine protein phosphatase-related
Length=476
Score = 60.1 bits (144), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 42/135 (31%), Positives = 69/135 (51%), Gaps = 8/135 (5%)
Query 132 SKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEA 191
S KE E + N+A++ Y++++ + P A+ Y RA K KRY A DC EA
Sbjct 85 SSEKEQGNEFFKQKKFNEAIDCYSRSIALS-PNAVTYANRAMAYLKIKRYREAEVDCTEA 143
Query 192 LKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKI-----DYDENIWDIQKLVEQKYK 246
L L+ +AY R T+ + LG K+A D E ++ + + DI+ L+E+ +
Sbjct 144 LNLDDRYIKAYSRRATARKELGMIKEAKEDAEFALRLEPESQELKKQYADIKSLLEK--E 201
Query 247 IIEEHERSVQRRREE 261
IIE+ ++Q +E
Sbjct 202 IIEKATGAMQSTAQE 216
> dre:768178 zgc:153288
Length=591
Score = 59.7 bits (143), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 58/96 (60%), Gaps = 1/96 (1%)
Query 133 KAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEAL 192
+ K+ EA + D +A++ Y ++L + + +A ++ RA+ L + +++ AA+ DCD L
Sbjct 196 REKDKGNEAYRSRDYEEALDYYCRSLSLAS-SAAVFNNRAQTLIRLQQWPAALSDCDAVL 254
Query 193 KLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKI 228
+L P N +A R T +++LG+ +++H D+ +I
Sbjct 255 QLEPHNIKALLRRATVHKHLGHQQESHDDLRAVLQI 290
Score = 59.7 bits (143), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 58/105 (55%), Gaps = 2/105 (1%)
Query 149 KAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTS 208
+ E +T++L + T YT RA K +R+ A +DCD AL++ P N +A+ R +
Sbjct 468 ETAERHTQSLQLDTHTCAAYTNRALCYIKLERFTEARQDCDSALQIEPTNKKAFYRRALA 527
Query 209 NRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHER 253
N+ L ++ SD++ Q + D ++ + Q+L+ + ++E+ R
Sbjct 528 NKGLKDYLSCRSDLQ--QVLRLDASVTEAQRLLMELTHLMEDRRR 570
> hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associated
antigen 1
Length=926
Score = 58.9 bits (141), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 49/197 (24%), Positives = 91/197 (46%), Gaps = 11/197 (5%)
Query 76 KIPDPLAEEPKEESEEEPFHVESLNDEEVVAPESTPPPALAPAGEKELTDDDYEKL-SKA 134
K P AE K + E+E ++ E+ V +S LT+ + + L ++
Sbjct 153 KTPRDYAEWDKFDVEKECLKIDEDYKEKTVIDKSHLSKIETRIDTAGLTEKEKDFLATRE 212
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
KE EA +GD +AV YT+++ PT + Y RA+ K + + +A +DC++ L+L
Sbjct 213 KEKGNEAFNSGDYEEAVMYYTRSI-SALPTVVAYNNRAQAEIKLQNWNSAFQDCEKVLEL 271
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERS 254
P N +A R T+ ++ ++A D+ + D++ + K + E ER
Sbjct 272 EPGNVKALLRRATTYKHQNKLREATEDL---------SKVLDVEPDNDLAKKTLSEVERD 322
Query 255 VQRRREEKERREREKRI 271
++ E + + KR+
Sbjct 323 LKNSEAASETQTKGKRM 339
Score = 57.8 bits (138), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 64/262 (24%), Positives = 113/262 (43%), Gaps = 45/262 (17%)
Query 106 APESTPPPALAPAGE--------------KELTDDDYEKLSKA-KEAAAEALEAGDLNKA 150
P S P A PA E + +TD EK KA KE + + + A
Sbjct 586 VPASVPLQAWHPAKEMISKQAGDSSSHRQQGITD---EKTFKALKEEGNQCVNDKNYKDA 642
Query 151 VESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNR 210
+ Y++ L + N +YT RA K ++ A +DCD+AL+L N +A+ R +++
Sbjct 643 LSKYSECLKINNKECAIYTNRALCYLKLCQFEEAKQDCDQALQLADGNVKAFYRRALAHK 702
Query 211 YLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERSVQRRREEKERREREKR 270
L N+++ S I++ + I D +I + + +E+ +++ +++ + EKERR+ E
Sbjct 703 GLKNYQK--SLIDLNKVILLDPSIIEAKMELEEVTRLLNLKDKTAPFNK-EKERRKIE-- 757
Query 271 IREQRASSQKAYEEQKKREASGVPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIP 330
I+E ++ PG GE+S G GG S P P
Sbjct 758 IQEVNEGKEE---------------------PGRPAGEVSMGCLASEKGGKSSRSPED-P 795
Query 331 GGVPGGMPGGMGGLGGLLGSVN 352
+P P G ++ +++
Sbjct 796 EKLPIAKPNNAYEFGQIINALS 817
Score = 37.7 bits (86), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 23/98 (23%), Positives = 46/98 (46%), Gaps = 8/98 (8%)
Query 140 EALEAGDLNKAVESYTKALLVGNPT--------ALLYTRRAEVLFKQKRYAAAVRDCDEA 191
E +G +A Y+ A+ + P ++LY+ RA K+ + ++DC+ A
Sbjct 454 ELFRSGQFAEAAGKYSAAIALLEPAGSEIADDLSILYSNRAACYLKEGNCSGCIQDCNRA 513
Query 192 LKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID 229
L+L+P + + R + L + +A+ D + +ID
Sbjct 514 LELHPFSMKPLLRRAMAYETLEQYGKAYVDYKTVLQID 551
> dre:564953 spag1, MGC162178, cb1089, wu:fj78g10; sperm associated
antigen 1
Length=386
Score = 58.2 bits (139), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 68/137 (49%), Gaps = 2/137 (1%)
Query 114 ALAPAGEKELTDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAE 173
A A E+E + + K+ E ++ A E Y++ L + +YT RA
Sbjct 244 ARAERAEQEKARKAEARFTILKQEGNELVKNSQFQGASEKYSECLAIKPNECAIYTNRAL 303
Query 174 VLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDEN 233
K +R+A A +DCD AL++ P N +A+ R +++ L ++ A +D++ + + D N
Sbjct 304 CFLKLERFAEAKQDCDSALQMEPKNKKAFYRRALAHKGLKDYLSASTDLQ--EVLQLDPN 361
Query 234 IWDIQKLVEQKYKIIEE 250
+ + ++ +E ++ E
Sbjct 362 VQEAEQELEMVTNLLRE 378
Score = 41.2 bits (95), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 59/127 (46%), Gaps = 14/127 (11%)
Query 131 LSKAKEAAAEALEAGDLNKAVESYTKALL------VGNPTAL--LYTRRAEVLFKQKRYA 182
L++ K + G A+E YT+A+ + +P L LY+ RA K A
Sbjct 84 LARLKNQGNMLFKNGQFGDALEKYTQAIDGCIEAGIDSPEDLCVLYSNRAACFLKDGNSA 143
Query 183 AAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYD-----ENIWDI 237
++DC AL+L+P + + R + L +++A+ D + +ID +++ I
Sbjct 144 DCIQDCTRALELHPFSLKPLLRRAMAYESLERYRKAYVDYKTVLQIDISVQAAHDSVHRI 203
Query 238 QK-LVEQ 243
K L+EQ
Sbjct 204 TKMLIEQ 210
> cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);
K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=496
Score = 57.0 bits (136), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 62/109 (56%), Gaps = 1/109 (0%)
Query 120 EKELTDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQK 179
E++ +D+ EK K+ A + + + A + Y+ A+ + +PTA+LY RA+ K++
Sbjct 18 EEKSYEDEKEKAGMIKDEANQFFKDQVYDVAADLYSVAIEI-HPTAVLYGNRAQAYLKKE 76
Query 180 RYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKI 228
Y +A+ D D A+ ++P + + R T+N LG +K+A +D + K+
Sbjct 77 LYGSALEDADNAIAIDPSYVKGFYRRATANMALGRFKKALTDYQAVVKV 125
> xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subunit
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=493
Score = 56.2 bits (134), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 58/130 (44%), Gaps = 9/130 (6%)
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
KE A E D + AV+ YT+A+ + TA+ Y R+ + + Y A+ D A++L
Sbjct 26 KEQANEYFRVKDYDHAVQYYTQAIDLSPDTAIYYGNRSLAYLRTECYGYALADASRAIQL 85
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKI-----DYDENIWDIQKLVEQK----Y 245
+ + Y R SN LG K A D E K+ D + KLV QK
Sbjct 86 DAKYIKGYYRRAASNMALGKLKAALKDYETVVKVRPHDKDAQMKFQECNKLVRQKAFERA 145
Query 246 KIIEEHERSV 255
E+H RSV
Sbjct 146 IACEQHNRSV 155
> xla:496358 sgta; small glutamine-rich tetratricopeptide repeat
(TPR)-containing, alpha
Length=302
Score = 55.8 bits (133), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 59/124 (47%), Gaps = 1/124 (0%)
Query 124 TDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAA 183
+D+D + + K E ++ + A+ YTKAL + A+ Y RA K YA
Sbjct 69 SDEDLAEAERLKTEGNEQMKVENFESAISYYTKALELNPANAVYYCNRAAAYSKLGNYAG 128
Query 184 AVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQ-KLVE 242
AVRDC+ A+ ++P+ ++AY G + L +A + +D D + K+ E
Sbjct 129 AVRDCEAAITIDPNYSKAYGRMGLALSSLNKHAEAVGFYKQALVLDPDNETYKSNLKIAE 188
Query 243 QKYK 246
QK K
Sbjct 189 QKMK 192
> ath:AT1G33400 tetratricopeptide repeat (TPR)-containing protein
Length=798
Score = 55.1 bits (131), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 39/126 (30%), Positives = 59/126 (46%), Gaps = 15/126 (11%)
Query 113 PALAPAGEKELTDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLV-------GNPTA 165
P L G+ E T D K + D ++A+ Y+KAL V G+ +
Sbjct 51 PELGCCGKNEETSLDL------KRRGNHCFRSRDFDEALRLYSKALRVAPLDAIDGDKSL 104
Query 166 L--LYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIE 223
L L+ RA VL ++RDC AL+++P A+A+ RG N LGN+K A DI
Sbjct 105 LASLFLNRANVLHNLGLLKESLRDCHRALRIDPYYAKAWYRRGKLNTLLGNYKDAFRDIT 164
Query 224 MGQKID 229
+ ++
Sbjct 165 VSMSLE 170
> dre:323361 tomm34, wu:fb96b08, zgc:56645; translocase of outer
mitochondrial membrane 34
Length=305
Score = 55.1 bits (131), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 57/113 (50%), Gaps = 2/113 (1%)
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
KE ++ G+ KA+E YT++L YT RA K Y A+RDC+EAL+L
Sbjct 194 KEEGNALVKKGEHKKAMEKYTQSLAQDPTEVTTYTNRALCYLALKMYKDAIRDCEEALRL 253
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKI 247
+ N +A R + + L N K D+ KI D N +QKL+++ K+
Sbjct 254 DSANIKALYRRAQAYKELKNKKSCIEDLNSVLKI--DPNNTAVQKLLQEVQKM 304
Score = 37.7 bits (86), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 48/105 (45%), Gaps = 8/105 (7%)
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTA--------LLYTRRAEVLFKQKRYAAAVR 186
K+A E +AG +AV Y++A+ + +LY+ RA K ++
Sbjct 14 KQAGNECFKAGQYGEAVTLYSQAIQQLEKSGQKKTEDLGILYSNRAASYLKDGNCNECIK 73
Query 187 DCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYD 231
DC +L L P +A R + L ++QA+ D + +ID++
Sbjct 74 DCTASLDLVPFGFKALLRRAAAFEALERYRQAYVDYKTVLQIDWN 118
> mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase
II associated protein 3
Length=660
Score = 54.7 bits (130), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 145 GDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRI 204
G +A+E YT+ + ALL RA K +RY A RDC +A+ L+ ++A+
Sbjct 298 GKYEQAIECYTRGIAADRTNALLPANRAMAYLKIQRYEEAERDCTQAIVLDGSYSKAFAR 357
Query 205 RGTSNRYLGNWKQAHSDIE 223
RGT+ +LG +A D E
Sbjct 358 RGTARTFLGKINEAKQDFE 376
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 60/143 (41%), Gaps = 19/143 (13%)
Query 81 LAEEPKEESEEEPFHVESLNDEEVVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAAAE 140
L E KE+S + ES +DE+ + +S L G K
Sbjct 103 LDELDKEDSTHDSLSQESESDEDGIRVDSQKALVLKEKGNK------------------- 143
Query 141 ALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNAR 200
+ G ++A+E YTK + +L T RA F+ K++A A DC+ A+ L+ +
Sbjct 144 YFKQGKYDEAIECYTKGMDADPYNPVLPTNRASAYFRLKKFAVAESDCNLAIALSRTYTK 203
Query 201 AYRIRGTSNRYLGNWKQAHSDIE 223
AY RG + L + A D E
Sbjct 204 AYARRGAARFALQKLEDARKDYE 226
> xla:414520 hypothetical protein MGC81394
Length=312
Score = 54.3 bits (129), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 58/124 (46%), Gaps = 1/124 (0%)
Query 124 TDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAA 183
+D+D + K E ++ + AV YTKAL + A+ Y RA K YA
Sbjct 81 SDEDVAEAESLKTEGNEQMKVENFESAVTYYTKALELNPRNAVYYCNRAAAYSKLGNYAG 140
Query 184 AVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQ-KLVE 242
AVRDC+EA+ ++P ++AY G + L ++ + +D + + K+ E
Sbjct 141 AVRDCEEAISIDPSYSKAYGRMGLALSSLNKHAESVGFYKQALVLDPENETYKSNLKIAE 200
Query 243 QKYK 246
QK K
Sbjct 201 QKMK 204
> mmu:67145 Tomm34, 2610100K07Rik, TOM34, Tomm34a, Tomm34b; translocase
of outer mitochondrial membrane 34
Length=309
Score = 54.3 bits (129), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 51/90 (56%), Gaps = 0/90 (0%)
Query 127 DYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVR 186
D E+ KE + ++ G+ KA+E Y+++LL + + Y+ RA K+Y AV+
Sbjct 189 DVERAKALKEEGNDLVKKGNHKKAIEKYSESLLCSSLESATYSNRALCHLVLKQYKEAVK 248
Query 187 DCDEALKLNPDNARAYRIRGTSNRYLGNWK 216
DC EALKL+ N +A+ R + + L ++K
Sbjct 249 DCTEALKLDGKNVKAFYRRAQAYKALKDYK 278
> dre:563791 ttc12; tetratricopeptide repeat domain 12
Length=700
Score = 54.3 bits (129), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 46/90 (51%), Gaps = 0/90 (0%)
Query 129 EKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDC 188
E+ + +E EA GD AV YT+ L LYT RA+ K KRY A+ DC
Sbjct 111 ERANVLREQGNEAFTQGDYETAVRFYTEGLEQLRDMQALYTNRAQAFIKLKRYKEAISDC 170
Query 189 DEALKLNPDNARAYRIRGTSNRYLGNWKQA 218
+ AL+ N +A+ GTS+ L ++ Q+
Sbjct 171 EWALRCNEKCIKAFIHMGTSHLALKDFTQS 200
> sce:YOR027W STI1; Hsp90 cochaperone, interacts with the Ssa
group of the cytosolic Hsp70 chaperones; activates the ATPase
activity of Ssa1p; homolog of mammalian Hop protein; K09553
stress-induced-phosphoprotein 1
Length=589
Score = 53.9 bits (128), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 60/108 (55%), Gaps = 7/108 (6%)
Query 123 LTDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVG-NPTALLYTRRAEVLFKQKRY 181
LT D+Y++ A A A D +KA+E +TKA+ V P +LY+ R+ K++
Sbjct 3 LTADEYKQQGNA------AFTAKDYDKAIELFTKAIEVSETPNHVLYSNRSACYTSLKKF 56
Query 182 AAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID 229
+ A+ D +E +K+NP ++ Y G ++ LG+ +A S+ + ++D
Sbjct 57 SDALNDANECVKINPSWSKGYNRLGAAHLGLGDLDEAESNYKKALELD 104
Score = 38.1 bits (87), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 55/117 (47%), Gaps = 0/117 (0%)
Query 129 EKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDC 188
EK +A+ E D AV++YT+ + A Y+ RA L K + A+ DC
Sbjct 394 EKAEEARLEGKEYFTKSDWPNAVKAYTEMIKRAPEDARGYSNRAAALAKLMSFPEAIADC 453
Query 189 DEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKY 245
++A++ +P+ RAY + T+ + + A ++ + D + N + ++Q Y
Sbjct 454 NKAIEKDPNFVRAYIRKATAQIAVKEYASALETLDAARTKDAEVNNGSSAREIDQLY 510
> mmu:353282 Sfmbt2, D2Wsu23e, D330030P06Rik; Scm-like with four
mbt domains 2
Length=975
Score = 53.9 bits (128), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 34/61 (55%), Gaps = 0/61 (0%)
Query 281 AYEEQKKREASGVPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGG 340
Y +++++ A VP G P G+P G P ++ G P G P + S+P GIP +P +P
Sbjct 730 VYVQKRRKSAIVVPAGVPAGVPAGVPEDIPAGIPEGIPASIPESIPEGIPESLPESLPEA 789
Query 341 M 341
+
Sbjct 790 I 790
> ath:AT3G58620 TTL4; TTL4 (Tetratricopetide-repeat Thioredoxin-Like
4); binding
Length=682
Score = 52.0 bits (123), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 52/102 (50%), Gaps = 0/102 (0%)
Query 126 DDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAV 185
++ + ++KA+ E +G ++A +Y L + ++LY RA FK + +V
Sbjct 444 NNVKNVAKARTRGNELFSSGRYSEASVAYGDGLKLDAFNSVLYCNRAACWFKLGMWEKSV 503
Query 186 RDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQK 227
DC++AL++ P +A R S LG W+ A D E+ +K
Sbjct 504 DDCNQALRIQPSYTKALLRRAASYGKLGRWEDAVRDYEVLRK 545
Score = 31.6 bits (70), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 37/145 (25%), Positives = 65/145 (44%), Gaps = 23/145 (15%)
Query 130 KLSKAKEAAAEALEA-------------GDLNKAVESYTKALLVGNPTALLYTRRAEVLF 176
K+S A +AAAE ++ G+ +A+ Y +A+ + + RA L
Sbjct 197 KVSHATKAAAEMSDSEEVKKAGNVMYRKGNYAEALALYDRAISLSPENPAYRSNRAAALA 256
Query 177 KQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEM-GQKIDYDENIW 235
R AV++C EA++ +P ARA++ + LG + A + + GQ D
Sbjct 257 ASGRLEEAVKECLEAVRCDPSYARAHQRLASLYLRLGEAENARRHLCVSGQCPDQA---- 312
Query 236 DIQKLVEQKYKIIEEHERSVQRRRE 260
D+Q+L + +E+H R R+
Sbjct 313 DLQRL-----QTLEKHLRLCTEARK 332
> ath:AT5G09420 ATTOC64-V (ARABIDOPSIS THALIANA TRANSLOCON AT
THE OUTER MEMBRANE OF CHLOROPLASTS 64-V); amidase/ binding /
carbon-nitrogen ligase, with glutamine as amido-N-donor
Length=603
Score = 51.6 bits (122), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 52/108 (48%), Gaps = 3/108 (2%)
Query 115 LAPAGEKELTDDDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEV 174
LAP + T+ + E KE A + NKAV YT+A+ + A Y RA
Sbjct 475 LAPVSD---TNGNMEASEVMKEKGNAAYKGKQWNKAVNFYTEAIKLNGANATYYCNRAAA 531
Query 175 LFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDI 222
+ + A +DC +A+ ++ N +AY RGT+ L +K+A +D
Sbjct 532 FLELCCFQQAEQDCTKAMLIDKKNVKAYLRRGTARESLVRYKEAAADF 579
> dre:493606 stip1, zgc:92133; stress-induced-phosphoprotein 1
(Hsp70/Hsp90-organizing protein); K09553 stress-induced-phosphoprotein
1
Length=542
Score = 51.6 bits (122), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 54/97 (55%), Gaps = 0/97 (0%)
Query 129 EKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDC 188
EK+S+ K+ +AL AG+L +A+ YT+AL + +L++ R+ K+ Y A++D
Sbjct 2 EKVSQLKDQGNKALSAGNLEEAIRCYTEALTLDPSNHVLFSNRSAAYAKKGDYDNALKDA 61
Query 189 DEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMG 225
+ +K+ PD + Y + + +LG + A + + G
Sbjct 62 CQTIKIKPDWGKGYSRKAAALEFLGRLEDAKATYQEG 98
Score = 34.7 bits (78), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 350 SVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLM 396
++ DP+VQQ+ S+P M + + +P ++ + +P +A I KL+
Sbjct 488 AMADPEVQQIMSDPAMRMILEQMQKDPQALSDHLKNPVIAQKIQKLI 534
> hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein
3
Length=631
Score = 50.8 bits (120), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 145 GDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRI 204
G +A+E YT+ + ALL RA K ++Y A +DC +A+ L+ ++A+
Sbjct 296 GKYERAIECYTRGIAADGANALLPANRAMAYLKIQKYEEAEKDCTQAILLDGSYSKAFAR 355
Query 205 RGTSNRYLGNWKQAHSDIE 223
RGT+ +LG +A D E
Sbjct 356 RGTARTFLGKLNEAKQDFE 374
Score = 47.4 bits (111), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 48/97 (49%), Gaps = 0/97 (0%)
Query 127 DYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVR 186
D +K KE + + G ++A++ YTK + +L T RA F+ K++A A
Sbjct 129 DSQKALVLKEKGNKYFKQGKYDEAIDCYTKGMDADPYNPVLPTNRASAYFRLKKFAVAES 188
Query 187 DCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIE 223
DC+ A+ LN +AY RG + L ++A D E
Sbjct 189 DCNLAVALNRSYTKAYSRRGAARFALQKLEEAKKDYE 225
> dre:336867 fk20d10, wu:fa05b08, wu:fc52b05, wu:fk20d10, zgc:77080;
zgc:55741
Length=320
Score = 50.1 bits (118), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 55/102 (53%), Gaps = 1/102 (0%)
Query 146 DLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIR 205
+ + AVE Y+KA+ + A+ + RA K YA AV+DC+ A+ ++ + ++AY
Sbjct 106 NFSAAVEFYSKAIQLNPQNAVYFCNRAAAYSKLGNYAGAVQDCERAIGIDANYSKAYGRM 165
Query 206 GTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQ-KLVEQKYK 246
G + L + +A S + ++D D + + + ++ EQK K
Sbjct 166 GLALASLNKYSEAVSYYKKALELDPDNDTYKVNLQVAEQKVK 207
> dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, alpha
Length=306
Score = 49.7 bits (117), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/156 (25%), Positives = 71/156 (45%), Gaps = 13/156 (8%)
Query 92 EPFHVESLNDEEVVAPESTPPPALAPAGEKELTDDDYEKLSKAKEAAAEALEAGDLNKAV 151
E F L ++ V P++ P P +D E+ + K ++ + + AV
Sbjct 60 EIFLNSLLKNDIVTLPKTFPSP------------EDIERAEQLKNEGNNHMKEENYSSAV 107
Query 152 ESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRY 211
+ YTKA+ + A+ Y RA K + Y A+ DC+ A+ ++P ++AY G +
Sbjct 108 DCYTKAIELDQRNAVYYCNRAAAHSKLENYTEAMGDCERAIAIDPSYSKAYGRMGLALTS 167
Query 212 LGNWKQAHSDIEMGQKIDYDENIWDIQ-KLVEQKYK 246
+ + +A S +D + + + K+VEQK K
Sbjct 168 MSKYPEAISYFNKALVLDPENDTYKSNLKIVEQKQK 203
> dre:393173 MGC56178; zgc:56178
Length=595
Score = 48.9 bits (115), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 55/111 (49%), Gaps = 2/111 (1%)
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
KE + + G + A+E YTKA+ + T RA ++ K++A A DC+ A+ L
Sbjct 132 KEKGNQFFKDGRFDSAIECYTKAMDADPYNPVPPTNRATCFYRLKKFAVAESDCNLAIAL 191
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID--YDENIWDIQKLVEQ 243
+ +AY R + L ++A D EM K+D E ++QKL ++
Sbjct 192 DSKYVKAYIRRAATRTALQKHREALEDYEMVLKLDPGNSEAQTEVQKLQQE 242
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 48/94 (51%), Gaps = 2/94 (2%)
Query 150 AVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSN 209
AVESYT+ + ALL RA K R+A A +DC AL L+P +A+ R T+
Sbjct 298 AVESYTRGMEADETNALLPANRAMAFLKLNRFAEAEQDCSAALALDPSYTKAFARRATAR 357
Query 210 RYLGNWKQAHSDIEMGQKID--YDENIWDIQKLV 241
LG + A D E K++ + I +I+KL
Sbjct 358 AALGKCRDARDDFEQVLKLEPGNKQAISEIEKLT 391
> ath:AT4G11260 SGT1B; SGT1B; protein binding; K12795 suppressor
of G2 allele of SKP1
Length=358
Score = 48.5 bits (114), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 63/122 (51%), Gaps = 3/122 (2%)
Query 136 EAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLN 195
E A EA D + AV+ Y+KA+ + A + RA+ K + AV D ++A++L
Sbjct 7 EKAKEAFLDDDFDVAVDLYSKAIDLDPNCAAFFADRAQANIKIDNFTEAVVDANKAIELE 66
Query 196 PDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQ-KYKIIEEHERS 254
P A+AY +GT+ L + A + +E G + +E +K++++ +I EE +
Sbjct 67 PTLAKAYLRKGTACMKLEEYSTAKAALEKGASVAPNEP--KFKKMIDECDLRIAEEEKDL 124
Query 255 VQ 256
VQ
Sbjct 125 VQ 126
> tgo:TGME49_047000 TPR domain-containing protein (EC:3.4.21.72)
Length=888
Score = 48.1 bits (113), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 55/115 (47%), Gaps = 12/115 (10%)
Query 157 ALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWK 216
AL A+L + R K + A V DC EA++ +P A+AY R T+N L W
Sbjct 736 ALEFNQLRAVLLSNRGACHLHGKCWEAVVADCTEAIQCDPSYAKAYLRRFTANEALTKWH 795
Query 217 QAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERSVQRRREEKERREREKRI 271
A +DI ++D +E +Y+ ++ V+++ E + +E+E+ I
Sbjct 796 DAAADINKAIELDPS---------LEARYR---SDQQRVKKKSEAQFEKEKEEMI 838
> mmu:100048137 tetratricopeptide repeat protein 12-like
Length=544
Score = 48.1 bits (113), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
KE EA GD A+ Y++ L +LYT RA+ K Y A+ DCD ALK
Sbjct 109 KEKGNEAFVRGDYETAIFFYSEGLGKLKDMKVLYTNRAQAFIKLGDYQKALVDCDWALKC 168
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID 229
+ + +AY G ++ L N+ +A E QKI+
Sbjct 169 DENCTKAYFHMGKAHVALKNYSKAK---ECYQKIE 200
> ath:AT2G42580 TTL3; TTL3 (TETRATRICOPETIDE-REPEAT THIOREDOXIN-LIKE
3); binding / protein binding
Length=691
Score = 48.1 bits (113), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 48/102 (47%), Gaps = 0/102 (0%)
Query 126 DDYEKLSKAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAV 185
++ + + +A+ E +G ++A +Y L + ++LY RA +K + +V
Sbjct 453 NNVKMVVRARTRGNELFSSGRFSEACVAYGDGLKQDDSNSVLYCNRAACWYKLGLWEKSV 512
Query 186 RDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQK 227
DC+ ALK P +A R S LG W+ A D E ++
Sbjct 513 EDCNHALKSQPSYIKALLRRAASYGKLGRWEDAVKDYEFLRR 554
Score = 40.0 bits (92), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 57/117 (48%), Gaps = 10/117 (8%)
Query 145 GDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRI 204
G ++A+ Y +A+L+ A + RA L +R AV++C EA++++P +RA++
Sbjct 234 GSFSEALSLYDRAILISPGNAAYRSNRAAALTALRRLGEAVKECLEAVRIDPSYSRAHQR 293
Query 205 RGTSNRYLGNWKQAHSDIEM-GQKIDYDENIWDIQKLVEQKYKIIEEHERSVQRRRE 260
+ LG + A I GQ D D+Q+L + +E+H R R+
Sbjct 294 LASLYLRLGEAENARRHICFSGQCPDQA----DLQRL-----QTLEKHLRRCWEARK 341
> dre:568208 ttc6; tetratricopeptide repeat domain 6
Length=720
Score = 47.8 bits (112), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 44/79 (55%), Gaps = 0/79 (0%)
Query 151 VESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNR 210
V+ + +AL + ++ RA + + R+A A+ DC+EA+++ P + RA+ +G
Sbjct 260 VQDFNRALKINPKFYQVHLSRAALYGAEGRHAKAILDCNEAIRIQPKSLRAHLYKGALKF 319
Query 211 YLGNWKQAHSDIEMGQKID 229
YL +K A D+ M +ID
Sbjct 320 YLKAYKSAVEDLTMAVQID 338
Score = 33.5 bits (75), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 40/79 (50%), Gaps = 0/79 (0%)
Query 150 AVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSN 209
+++ + +AL + +A Y RA + +++ +A RD +AL L P +A Y++R
Sbjct 639 SLQDFNEALCLNPLSAHAYFNRANLHCSLRQFQSAERDLTQALVLEPGDALLYKLRADVR 698
Query 210 RYLGNWKQAHSDIEMGQKI 228
LG ++A D K+
Sbjct 699 GCLGWMEEAMEDYRTALKL 717
> mmu:235330 Ttc12, E330017O07Rik; tetratricopeptide repeat domain
12
Length=704
Score = 47.8 bits (112), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
KE EA GD A+ Y++ L +LYT RA+ K Y A+ DCD ALK
Sbjct 109 KEKGNEAFVRGDYETAIFFYSEGLGKLKDMKVLYTNRAQAFIKLGDYQKALVDCDWALKC 168
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID 229
+ + +AY G ++ L N+ +A E QKI+
Sbjct 169 DENCTKAYFHMGKAHVALKNYSKAK---ECYQKIE 200
> ath:AT3G17970 atToc64-III (Arabidopsis thaliana translocon at
the outer membrane of chloroplasts 64-III); binding / carbon-nitrogen
ligase, with glutamine as amido-N-donor
Length=589
Score = 47.8 bits (112), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 45/89 (50%), Gaps = 0/89 (0%)
Query 134 AKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALK 193
AKE +A + KA+ Y++A+ + + A Y+ RA + + A DC +A+
Sbjct 477 AKEKGNQAFKEKLWQKAIGLYSEAIKLSDNNATYYSNRAAAYLELGGFLQAEEDCTKAIT 536
Query 194 LNPDNARAYRIRGTSNRYLGNWKQAHSDI 222
L+ N +AY RGT+ LG+ K A D
Sbjct 537 LDKKNVKAYLRRGTAREMLGDCKGAIEDF 565
Score = 32.0 bits (71), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 0/54 (0%)
Query 176 FKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID 229
FK+K + A+ EA+KL+ +NA Y R + LG + QA D +D
Sbjct 485 FKEKLWQKAIGLYSEAIKLSDNNATYYSNRAAAYLELGGFLQAEEDCTKAITLD 538
> hsa:7265 TTC1, FLJ46404, TPR1; tetratricopeptide repeat domain
1
Length=292
Score = 47.4 bits (111), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 43/160 (26%), Positives = 76/160 (47%), Gaps = 26/160 (16%)
Query 120 EKELTDDDYEKL----SKAKEAAAEALEAGDLNKAVESYTKAL-----LVGNPTALLYTR 170
EK ++D++ +K ++ KE E + GD +A SY++AL ++L++
Sbjct 101 EKNMSDEEKQKRREESTRLKEEGNEQFKKGDYIEAESSYSRALEMCPSCFQKERSILFSN 160
Query 171 RAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID- 229
RA KQ + A+ DC +A++LNP RA R E+ +K D
Sbjct 161 RAAARMKQDKKEMAINDCSKAIQLNPSYIRAILRRA----------------ELYEKTDK 204
Query 230 YDENIWDIQKLVEQKYKIIEEHERSVQRRREEKERREREK 269
DE + D + ++E+ I + E ++ ++ +ER ER K
Sbjct 205 LDEALEDYKSILEKDPSIHQAREACMRLPKQIEERNERLK 244
> ath:AT4G37460 SRFR1; SRFR1 (SUPPRESSOR OF RPS4-RLD 1); protein
complex scaffold
Length=1052
Score = 47.4 bits (111), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 50/170 (29%), Positives = 80/170 (47%), Gaps = 17/170 (10%)
Query 102 EEVVAPESTPPPAL-----APAGEKELTDD--DYEKLSKAKEAAAEALE--------AGD 146
++V+ E T P AL A A ++EL D+ K ++ AA+EA + G+
Sbjct 321 DKVLKEEPTYPEALIGRGTAYAFQRELESAIADFTKAIQSNPAASEAWKRRGQARAALGE 380
Query 147 LNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNARAYRIRG 206
+AVE TKAL+ + + R V FK K + AAV+D LK DN AY G
Sbjct 381 YVEAVEDLTKALVFEPNSPDVLHERGIVNFKSKDFTAAVKDLSICLKQEKDNKSAYTYLG 440
Query 207 TSNRYLGNWKQAHSDIEMGQKIDYDENIWDIQKLVEQKYKIIEEHERSVQ 256
+ LG +K+A + I D N + + Q Y+ + +H ++++
Sbjct 441 LAFASLGEYKKAEE--AHLKSIQLDSNYLEAWLHLAQFYQELADHCKALE 488
> mmu:209683 Ttc28, 2310015L07Rik, 6030435N04, AI428795, AI851761,
BC002262, MGC7623; tetratricopeptide repeat domain 28
Length=2481
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 40/74 (54%), Gaps = 0/74 (0%)
Query 140 EALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNA 199
+A GD + A+ Y +AL V +LY+ R+ K ++Y A+ D +A LNP
Sbjct 61 QACHDGDFHTAIVLYNEALAVDPQNCILYSNRSAAYMKTQQYHKALDDAIKARLLNPKWP 120
Query 200 RAYRIRGTSNRYLG 213
+AY +G + +YLG
Sbjct 121 KAYFRQGVALQYLG 134
> xla:379955 stip1, MGC53256; stress-induced-phosphoprotein 1;
K09553 stress-induced-phosphoprotein 1
Length=543
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 50/91 (54%), Gaps = 0/91 (0%)
Query 135 KEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKL 194
KE +AL AG+L++AV+ YT+A+ + +LY+ R+ K+K + A+ D + ++L
Sbjct 8 KEKGNKALSAGNLDEAVKCYTEAIKLDPKNHVLYSNRSAAYAKKKEFTKALEDGSKTVEL 67
Query 195 NPDNARAYRIRGTSNRYLGNWKQAHSDIEMG 225
D + Y + + +L +++A E G
Sbjct 68 KADWGKGYSRKAAALEFLNRFEEAKKTYEEG 98
Score = 42.7 bits (99), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 48/96 (50%), Gaps = 0/96 (0%)
Query 134 AKEAAAEALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALK 193
AK E+ + GD +A++ Y++A+ A LY+ RA K + AV+DC+E ++
Sbjct 363 AKNKGNESFQKGDYPQAMKHYSEAIKRNPNDAKLYSNRAACYTKLLEFLLAVKDCEECIR 422
Query 194 LNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID 229
L P + Y + + + ++ +A + ++D
Sbjct 423 LEPSFIKGYTRKAAALEAMKDFTKAMDAYQKAMELD 458
Score = 36.2 bits (82), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 350 SVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLM 396
++ DP+VQQ+ S+P M + + +P ++ + +P +A I KLM
Sbjct 489 AMADPEVQQIMSDPAMRLILEQMQKDPQALSDHLKNPVIAQKIQKLM 535
Score = 33.9 bits (76), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 33/126 (26%), Positives = 62/126 (49%), Gaps = 6/126 (4%)
Query 133 KAKEAAAEALEAGDLNKAVESYTKALLVGNPTALLY-TRRAEVLFKQKRYAAAVRDCDEA 191
K KE EA + D A++ Y +A + +P + Y T +A V F+ Y+ C++A
Sbjct 227 KEKELGNEAYKKKDFETALKHYGQAREL-DPANMTYITNQAAVYFEMGDYSKCRELCEKA 285
Query 192 LKLNPDNARAYRIRGTSNRYLGN--WKQAHSD--IEMGQKIDYDENIWDIQKLVEQKYKI 247
+++ +N YR+ + +GN +K+ + I+ K + ++ K +Q KI
Sbjct 286 IEVGRENREDYRLIAKAYARIGNSYFKEEKNKEAIQFFNKSLAEHRTPEVLKKCQQAEKI 345
Query 248 IEEHER 253
++E ER
Sbjct 346 LKEQER 351
> ath:AT5G16620 TIC40; TIC40
Length=447
Score = 46.2 bits (108), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 32/53 (60%), Gaps = 1/53 (1%)
Query 353 DPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGGGMAG 405
+PDV F NP++ AA + NP N+ KY++D EV D +K+ + F GM G
Sbjct 396 NPDVAMAFQNPRVQAALMECSENPMNIMKYQNDKEVMDVFNKISQLF-PGMTG 447
> mmu:66827 Ttc1, 4833412C19Rik, C79328, TPR1; tetratricopeptide
repeat domain 1
Length=292
Score = 46.2 bits (108), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 67/144 (46%), Gaps = 22/144 (15%)
Query 132 SKAKEAAAEALEAGDLNKAVESYTKAL-----LVGNPTALLYTRRAEVLFKQKRYAAAVR 186
+K KE E + GD +A SY++AL ++L++ RA KQ + A+
Sbjct 117 AKLKEEGNERFKRGDYMEAESSYSQALQMCPACFQKDRSVLFSNRAAARMKQDKKETAIT 176
Query 187 DCDEALKLNPDNARAYRIRGTSNRYLGNWKQAHSDIEMGQKID-YDENIWDIQKLVEQKY 245
DC +A++LNP RA R E+ +K D DE + D + ++E+
Sbjct 177 DCSKAIQLNPTYIRAILRRA----------------ELYEKTDKLDEALEDYKSVLEKDP 220
Query 246 KIIEEHERSVQRRREEKERREREK 269
+ + E ++ ++ +ER ER K
Sbjct 221 SVHQAREACMRLPKQIEERNERLK 244
> hsa:23331 TTC28, KIAA1043; tetratricopeptide repeat domain 28
Length=2481
Score = 46.2 bits (108), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 40/74 (54%), Gaps = 0/74 (0%)
Query 140 EALEAGDLNKAVESYTKALLVGNPTALLYTRRAEVLFKQKRYAAAVRDCDEALKLNPDNA 199
+A GD + A+ Y +AL V +LY+ R+ K ++Y A+ D +A LNP
Sbjct 67 QACHDGDFHTAIVLYNEALAVDPQNCILYSNRSAAYMKIQQYDKALDDAIKARLLNPKWP 126
Query 200 RAYRIRGTSNRYLG 213
+AY +G + +YLG
Sbjct 127 KAYFRQGVALQYLG 140
Lambda K H
0.312 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 18693687008
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40