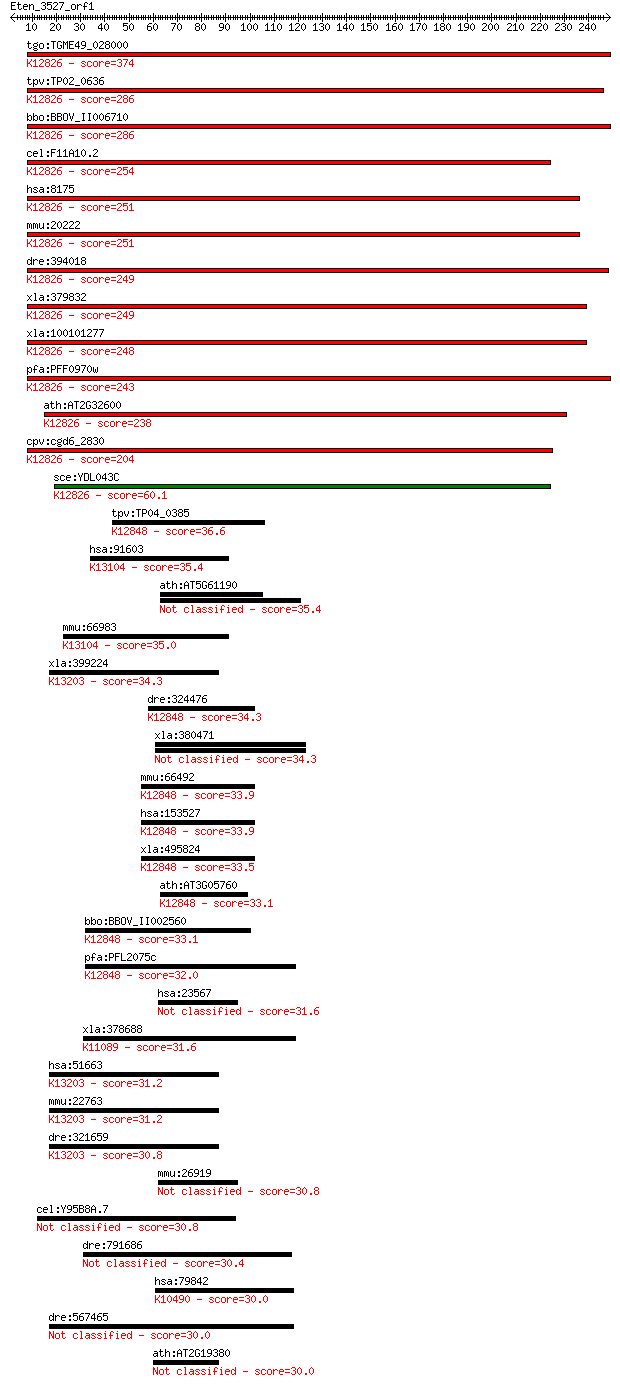
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3527_orf1
Length=248
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_028000 splicing factor 3A subunit 2, putative ; K12... 374 1e-103
tpv:TP02_0636 splicing factor 3A subunit 2; K12826 splicing fa... 286 4e-77
bbo:BBOV_II006710 18.m06549; splicing factor 3a, subunit 2; K1... 286 7e-77
cel:F11A10.2 hypothetical protein; K12826 splicing factor 3A s... 254 3e-67
hsa:8175 SF3A2, PRP11, PRPF11, SAP62, SF3a66; splicing factor ... 251 1e-66
mmu:20222 Sf3a2, 66kDa, AW047242, PRP11, SFA66, Sap62; splicin... 251 2e-66
dre:394018 sf3a2, MGC56097, zgc:56097, zgc:77448; splicing fac... 249 6e-66
xla:379832 sf3a2, MGC52805; splicing factor 3a, subunit 2, 66k... 249 8e-66
xla:100101277 hypothetical protein LOC100101277; K12826 splici... 248 1e-65
pfa:PFF0970w splicing factor 3a subunit, putative; K12826 spli... 243 5e-64
ath:AT2G32600 hydroxyproline-rich glycoprotein family protein;... 238 2e-62
cpv:cgd6_2830 splicing factor 3a 66kD; N-terminus C2H2 domain ... 204 3e-52
sce:YDL043C PRP11, RNA11; Prp11p; K12826 splicing factor 3A su... 60.1 6e-09
tpv:TP04_0385 hypothetical protein; K12848 U4/U6.U5 tri-snRNP ... 36.6 0.076
hsa:91603 ZNF830, CCDC16, MGC20398, OMCG1, SEL13; zinc finger ... 35.4 0.18
ath:AT5G61190 zinc finger protein-related 35.4 0.22
mmu:66983 Zfp830, 2410003C20Rik, AV301118, AW048207, Ccdc16, O... 35.0 0.25
xla:399224 zfr; zinc finger RNA binding protein; K13203 zinc f... 34.3 0.40
dre:324476 zmat2, fc30d10, flj31121, wu:fc30d10; zinc finger, ... 34.3 0.46
xla:380471 znf346, DsRBP-ZFa, MGC52555, ZFa, jaz; zinc finger ... 34.3 0.47
mmu:66492 Zmat2, 2610510D14Rik, 2900082I05Rik; zinc finger, ma... 33.9 0.53
hsa:153527 ZMAT2, FLJ31121, Snu23, hSNU23; zinc finger, matrin... 33.9 0.53
xla:495824 zmat2, snu23; zinc finger, matrin-type 2; K12848 U4... 33.5 0.68
ath:AT3G05760 nucleic acid binding / zinc ion binding; K12848 ... 33.1 1.0
bbo:BBOV_II002560 18.m06206; hypothetical protein; K12848 U4/U... 33.1 1.0
pfa:PFL2075c conserved Plasmodium protein; K12848 U4/U6.U5 tri... 32.0 2.2
hsa:23567 ZNF346, DKFZp547M223, JAZ, Zfp346; zinc finger prote... 31.6 2.5
xla:378688 trove2, ssa2, ssa2-A; TROVE domain family, member 2... 31.6 3.0
hsa:51663 ZFR, FLJ41312, ZFR1; zinc finger RNA binding protein... 31.2 3.4
mmu:22763 Zfr, C920030H05Rik, MGC176100; zinc finger RNA bindi... 31.2 3.5
dre:321659 zfr, hnrpa1, wu:fb25b08, zgc:66235, zgc:66403; zinc... 30.8 4.2
mmu:26919 Zfp346, Jaz, Znf346; zinc finger protein 346 30.8
cel:Y95B8A.7 hypothetical protein 30.8 5.1
dre:791686 cadherin 1, type 1 preproprotein-like 30.4 6.0
hsa:79842 ZBTB3, FLJ23392; zinc finger and BTB domain containi... 30.0 7.2
dre:567465 zfr2, MGC174987, si:ch211-63j24.3, wu:fb99a09, zfr;... 30.0 7.8
ath:AT2G19380 RNA recognition motif (RRM)-containing protein 30.0 8.5
> tgo:TGME49_028000 splicing factor 3A subunit 2, putative ; K12826
splicing factor 3A subunit 2
Length=242
Score = 374 bits (961), Expect = 1e-103, Method: Compositional matrix adjust.
Identities = 177/241 (73%), Positives = 204/241 (84%), Gaps = 2/241 (0%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
+DYQNRVGHK GSGAPAT QE NLERKERLRRLALET+DLNKDPYFM+NHLG FECRLCL
Sbjct 4 IDYQNRVGHKTGSGAPATAQEWNLERKERLRRLALETVDLNKDPYFMKNHLGHFECRLCL 63
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NEGSYL+HTQG+KHQTNL+RR KEKA++ VAP+P K + P R V+IGRPGY
Sbjct 64 TLHVNEGSYLAHTQGRKHQTNLARRKEKEKAESAVAPVPAKVA--PSRVGFTVKIGRPGY 121
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
RV+KLRD + Q ALL E++YPEI G KPYHRFMS+FEQRVE+PPD YQFLLFAA+PY
Sbjct 122 RVSKLRDPDSLQKALLFEIDYPEINEGAKPYHRFMSSFEQRVESPPDTKYQFLLFAADPY 181
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRPLPALPQRPTTFLPVGAR 247
ETIAFKIPNME+DR + + ++ WD EKKVYT+Q+FF KR + LPALPQRPT+FLPV AR
Sbjct 182 ETIAFKIPNMEVDRSEGKFYSNWDAEKKVYTIQLFFLKRSVKALPALPQRPTSFLPVTAR 241
Query 248 W 248
W
Sbjct 242 W 242
> tpv:TP02_0636 splicing factor 3A subunit 2; K12826 splicing
factor 3A subunit 2
Length=252
Score = 286 bits (732), Expect = 4e-77, Method: Compositional matrix adjust.
Identities = 141/239 (58%), Positives = 182/239 (76%), Gaps = 5/239 (2%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
+DYQ+RVGHK GSGAPA+ +V ++ERLR+LALET DLNKDPYF +NH+G ECRLCL
Sbjct 2 IDYQHRVGHKTGSGAPASAHDVAAHQRERLRKLALETFDLNKDPYFSKNHMGHVECRLCL 61
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
T+HT E SYLSHTQG+KHQ NL+RR AKE+ DA + P+ + T ++IGRPGY
Sbjct 62 TVHTTESSYLSHTQGRKHQMNLARRAAKEQKDAFMTIAPISKTRAFKAPT--LKIGRPGY 119
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
R+TK+RD T+Q ALL E+E+PEI +GT P +RFMSAFEQ++E PPD NYQFLLFAA+PY
Sbjct 120 RITKMRDPETKQPALLFEIEFPEI-QGT-PKYRFMSAFEQKIEIPPDPNYQFLLFAADPY 177
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFS-KRQPRPLPALPQRPTTFLPVG 245
ETIAFK+PN+EID + + +D+++K++ Q+ F K+ + LP LPQRPT F VG
Sbjct 178 ETIAFKVPNLEIDNGPNKLFSYFDDKRKLFIFQVHFKLKKTFKVLPGLPQRPTKFDHVG 236
> bbo:BBOV_II006710 18.m06549; splicing factor 3a, subunit 2;
K12826 splicing factor 3A subunit 2
Length=238
Score = 286 bits (731), Expect = 7e-77, Method: Compositional matrix adjust.
Identities = 141/242 (58%), Positives = 176/242 (72%), Gaps = 6/242 (2%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MDYQNRVGHK GSG PA+ Q++ ++RLRRLALET DL KDPY +NHLGQ ECRLCL
Sbjct 1 MDYQNRVGHKTGSGGPASAQDLASHNRDRLRRLALETFDLGKDPYLQKNHLGQLECRLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
T+H ++GSYLSHTQG+KHQ NL+RR AKE+ + P+P R T V+IGRPGY
Sbjct 61 TIHASDGSYLSHTQGRKHQMNLARRAAKEERNNFHQPIPKPVGM---RHTSTVKIGRPGY 117
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
R+TK+RD T QFALL E+EYP+I KP +R MSAFEQR+E PPD +QFLLFAA+PY
Sbjct 118 RITKMRDPETNQFALLFEIEYPDIE--GKPRYRVMSAFEQRMERPPDPAFQFLLFAAQPY 175
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQI-FFSKRQPRPLPALPQRPTTFLPVGA 246
ETI FK+PN+EID + + WDE K+Y +QI F + R + LPALP RP+ + VG
Sbjct 176 ETIGFKVPNLEIDDSPGKLYVHWDETLKLYVLQIQFRNSRTTKRLPALPHRPSVYTGVGP 235
Query 247 RW 248
++
Sbjct 236 KF 237
> cel:F11A10.2 hypothetical protein; K12826 splicing factor 3A
subunit 2
Length=222
Score = 254 bits (648), Expect = 3e-67, Method: Compositional matrix adjust.
Identities = 121/216 (56%), Positives = 160/216 (74%), Gaps = 5/216 (2%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MD+QNR G K GSG A+ + ++R+ERLR+LALETIDL KDPYFMRNH+G +EC+LCL
Sbjct 1 MDFQNRAGGKTGSGGVASAADAGVDRRERLRQLALETIDLQKDPYFMRNHIGTYECKLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NEGSYL+HTQGKKHQ NL+RR AKE+++ P P K + + V+IGRPGY
Sbjct 61 TLHNNEGSYLAHTQGKKHQANLARRAAKEQSEQPFLPAPQKAAV---ETKKFVKIGRPGY 117
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
+VTK RD Q ALL +++YPEI G P HRFMSA+EQ+++ PPD +Q+LLFAAEPY
Sbjct 118 KVTKERDPGAGQQALLFQIDYPEIADGIAPRHRFMSAYEQKIQ-PPDKRWQYLLFAAEPY 176
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFF 223
ETI FKIP+ E+D+ + + W+++ K + +Q+ F
Sbjct 177 ETIGFKIPSREVDKSE-KFWTMWNKDTKQFFLQVAF 211
> hsa:8175 SF3A2, PRP11, PRPF11, SAP62, SF3a66; splicing factor
3a, subunit 2, 66kDa; K12826 splicing factor 3A subunit 2
Length=464
Score = 251 bits (642), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 130/228 (57%), Positives = 163/228 (71%), Gaps = 4/228 (1%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MD+Q+R G K GSG A+ E N +R+ERLR+LALETID+NKDPYFM+NHLG +EC+LCL
Sbjct 1 MDFQHRPGGKTGSGGVASSSESNRDRRERLRQLALETIDINKDPYFMKNHLGSYECKLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NEGSYL+HTQGKKHQTNL+RR AKE +A P P K V+IGRPGY
Sbjct 61 TLHNNEGSYLAHTQGKKHQTNLARRAAKEAKEAPAQPAPEKVKV---EVKKFVKIGRPGY 117
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
+VTK RD Q +LL +++YPEI G P HRFMSA+EQR+E PPD +Q+LL AAEPY
Sbjct 118 KVTKQRDSEMGQQSLLFQIDYPEIAEGIMPRHRFMSAYEQRIE-PPDRRWQYLLMAAEPY 176
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRPLPALP 235
ETIAFK+P+ EID+ + + W+ E K + +Q F +P P+LP
Sbjct 177 ETIAFKVPSREIDKAEGKFWTHWNRETKQFFLQFHFKMEKPPAPPSLP 224
> mmu:20222 Sf3a2, 66kDa, AW047242, PRP11, SFA66, Sap62; splicing
factor 3a, subunit 2; K12826 splicing factor 3A subunit
2
Length=485
Score = 251 bits (641), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 130/228 (57%), Positives = 163/228 (71%), Gaps = 4/228 (1%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MD+Q+R G K GSG A+ E N +R+ERLR+LALETID+NKDPYFM+NHLG +EC+LCL
Sbjct 1 MDFQHRPGGKTGSGGVASSSESNRDRRERLRQLALETIDINKDPYFMKNHLGSYECKLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NEGSYL+HTQGKKHQTNL+RR AKE +A P P K V+IGRPGY
Sbjct 61 TLHNNEGSYLAHTQGKKHQTNLARRAAKEAKEAPAQPAPEKVKV---EVKKFVKIGRPGY 117
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
+VTK RD Q +LL +++YPEI G P HRFMSA+EQR+E PPD +Q+LL AAEPY
Sbjct 118 KVTKQRDTEMGQQSLLFQIDYPEIAEGIMPRHRFMSAYEQRIE-PPDRRWQYLLMAAEPY 176
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRPLPALP 235
ETIAFK+P+ EID+ + + W+ E K + +Q F +P P+LP
Sbjct 177 ETIAFKVPSREIDKAEGKFWTHWNRETKQFFLQFHFKMEKPPAPPSLP 224
> dre:394018 sf3a2, MGC56097, zgc:56097, zgc:77448; splicing factor
3a, subunit 2; K12826 splicing factor 3A subunit 2
Length=278
Score = 249 bits (636), Expect = 6e-66, Method: Compositional matrix adjust.
Identities = 132/240 (55%), Positives = 166/240 (69%), Gaps = 11/240 (4%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MD+Q+R G K GSG A+ E N +R+ERLR+LALETID+NKDPYFM+NHLG +EC+LCL
Sbjct 1 MDFQHRAGGKTGSGGVASASESNRDRRERLRQLALETIDINKDPYFMKNHLGSYECKLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NEGSYL+HTQGKKHQ+NL+RR AKE +A P P K V+IGRPGY
Sbjct 61 TLHNNEGSYLAHTQGKKHQSNLARRAAKEAKEAPAQPAPEKVKV---EVKKFVKIGRPGY 117
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
+VTK RD Q +LL +++YPEI G P HRFMSA+EQR+E PPD +Q+LLFAAEPY
Sbjct 118 KVTKQRDPEIGQQSLLFQIDYPEIAEGIGPRHRFMSAYEQRIE-PPDRRWQYLLFAAEPY 176
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRPLPALPQRPTTFLPVGAR 247
ETIAFK+P+ EID+ + R W+ E K + +Q F + P+ P PVG +
Sbjct 177 ETIAFKVPSREIDKAETRFWTHWNRETKQFFLQFHFKMEKALVAPSGP-------PVGVK 229
> xla:379832 sf3a2, MGC52805; splicing factor 3a, subunit 2, 66kDa;
K12826 splicing factor 3A subunit 2
Length=405
Score = 249 bits (635), Expect = 8e-66, Method: Compositional matrix adjust.
Identities = 130/231 (56%), Positives = 162/231 (70%), Gaps = 4/231 (1%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MD+Q+R G K GSG A+ E N +R+ERLR+LALETID+NKDPYFM+NHLG +EC+LCL
Sbjct 1 MDFQHRAGGKTGSGGVASASESNRDRRERLRQLALETIDINKDPYFMKNHLGSYECKLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NEGSYL+HTQGKKHQTNL+RR AKE +A P P K V+IGRPGY
Sbjct 61 TLHNNEGSYLAHTQGKKHQTNLARRAAKEAKEAPAQPAPEKVKV---EVKKFVKIGRPGY 117
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
+VTK RD Q +LL +++YPEI P HRFMSA+EQR+E PPD +Q+LL AAEPY
Sbjct 118 KVTKQRDPEMGQQSLLFQIDYPEIAESIMPRHRFMSAYEQRIE-PPDRRWQYLLMAAEPY 176
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRPLPALPQRP 238
ETIAFK+P+ EID+ + + W+ E K + +Q F +P P LP P
Sbjct 177 ETIAFKVPSREIDKVEGKFWTHWNRETKQFFLQFHFKTEKPPQAPTLPPAP 227
> xla:100101277 hypothetical protein LOC100101277; K12826 splicing
factor 3A subunit 2
Length=405
Score = 248 bits (633), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 129/231 (55%), Positives = 162/231 (70%), Gaps = 4/231 (1%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MD+Q+R G K GSG A+ E N +R+ERLR+LALETID+NKDPYFM+NHLG +EC+LCL
Sbjct 1 MDFQHRAGGKTGSGGVASSSESNRDRRERLRQLALETIDINKDPYFMKNHLGSYECKLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NEGSYL+HTQGKKHQTNL+RR AKE +A P P K V+IGRPGY
Sbjct 61 TLHNNEGSYLAHTQGKKHQTNLARRAAKEAKEAPAQPAPEKVKV---EVKKFVKIGRPGY 117
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
+VTK RD Q +LL +++YPEI P HRFMSA+EQR+E PPD +Q+LL AAEPY
Sbjct 118 KVTKQRDPEMAQQSLLFQIDYPEIAESIMPRHRFMSAYEQRIE-PPDRRWQYLLMAAEPY 176
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRPLPALPQRP 238
ETIAFK+P+ EID+ + + W+ E K + +Q F +P P +P P
Sbjct 177 ETIAFKVPSREIDKVEGKFWTHWNRETKQFFLQFHFKMEKPPTAPTMPLAP 227
> pfa:PFF0970w splicing factor 3a subunit, putative; K12826 splicing
factor 3A subunit 2
Length=233
Score = 243 bits (620), Expect = 5e-64, Method: Compositional matrix adjust.
Identities = 123/241 (51%), Positives = 168/241 (69%), Gaps = 8/241 (3%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MD+QNRVGHK GSG P + +++N ER+ERL++LALE ID+ KDPY ++N++G FEC+LCL
Sbjct 1 MDFQNRVGHKTGSGMPLSREDINQERRERLKQLALENIDITKDPYILKNNVGMFECKLCL 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGY 127
TLH NE SYL HTQGKKHQ NL++RL KEK + + K S P + V+IG+P Y
Sbjct 61 TLHNNESSYLCHTQGKKHQMNLAQRLLKEKNEMTTNRLN-KPVSEPRKI---VKIGKPRY 116
Query 128 RVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPY 187
VT++R+++ Q +L E+ +P I TKP RFMS+FEQ++E PD YQ+LLFAAEPY
Sbjct 117 DVTRVRNKKN-QLGILFELSFPNIKENTKPKFRFMSSFEQKIEA-PDKKYQYLLFAAEPY 174
Query 188 ETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRPLPALPQRPTTFLPVGAR 247
ETIAFKIPN++ID ++ ++ W ++KK++ MQI F P P + + FL R
Sbjct 175 ETIAFKIPNIDIDENEGFYY-KWFDKKKIFVMQIHFQNAFP-PGHVNLREHSNFLSSTNR 232
Query 248 W 248
W
Sbjct 233 W 233
> ath:AT2G32600 hydroxyproline-rich glycoprotein family protein;
K12826 splicing factor 3A subunit 2
Length=277
Score = 238 bits (607), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 124/216 (57%), Positives = 162/216 (75%), Gaps = 4/216 (1%)
Query 15 GHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCLTLHTNEG 74
G K GSG A+ Q ++R+ERLRRLALETIDL KDPYFMRNHLG +EC+LCLTLH NEG
Sbjct 6 GSKPGSGGAASGQNEAIDRRERLRRLALETIDLAKDPYFMRNHLGSYECKLCLTLHNNEG 65
Query 75 SYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRVRIGRPGYRVTKLRD 134
+YL+HTQGK+HQTNL++R A+E DA P PLK + R+ V+IGRPGYRVTK D
Sbjct 66 NYLAHTQGKRHQTNLAKRAAREAKDAPTKPQPLKRNVSVRRT---VKIGRPGYRVTKQYD 122
Query 135 ERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAEPYETIAFKI 194
+Q +LL ++EYPEI KP HRFMS++EQ+V+ P D +YQ+LLFAAEPYE IAFK+
Sbjct 123 PELQQRSLLFQIEYPEIEDNIKPRHRFMSSYEQKVQ-PYDKSYQYLLFAAEPYEIIAFKV 181
Query 195 PNMEIDRDDPRHHAAWDEEKKVYTMQIFFSKRQPRP 230
P+ E+D+ P+ + WD + K++T+Q++F +P P
Sbjct 182 PSTEVDKSTPKFFSHWDPDSKMFTLQVYFKPTKPEP 217
> cpv:cgd6_2830 splicing factor 3a 66kD; N-terminus C2H2 domain
; K12826 splicing factor 3A subunit 2
Length=216
Score = 204 bits (518), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 115/219 (52%), Positives = 155/219 (70%), Gaps = 9/219 (4%)
Query 8 MDYQNRVGHKLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCL 67
MDY+NR GHK GSGA A+ Q++ +ER+ERLRRLALE+IDL+KDPY+M+NHLGQ ECRLC
Sbjct 1 MDYENRGGHKTGSGALASSQDIAIERRERLRRLALESIDLSKDPYYMKNHLGQVECRLCS 60
Query 68 TLHTNEGSYLSHTQGKKHQTNLSRRLAKEKA-DAVVAPMPLKTSSGPGRSTDRV-RIGRP 125
T+HTNEGSYLSHTQG+KHQTNL+ R +KEK AVV P + P ++ R RIG+P
Sbjct 61 TIHTNEGSYLSHTQGRKHQTNLAYRASKEKNLKAVVKPQ----AENPEQAKPRAPRIGQP 116
Query 126 GYRVTKLRDERTRQFALLCEVEYPEIVRGTKPYHRFMSAFEQRVETPPDGNYQFLLFAAE 185
Y+V+K R+ T + C+ + EI+ P +R MS +EQ+VE P+ YQ+L AE
Sbjct 117 KYKVSKHREGSTGTNCVYCKFYFQEILEDHIPGYRIMSCWEQKVEK-PNPKYQYLFVGAE 175
Query 186 PYETIAFKIPNMEIDRDDPRHHAAWDEEKKVYTMQIFFS 224
PY TI +IPN+E+ + R WDE++K+Y +Q++ S
Sbjct 176 PYNTIGIRIPNIELIKQ--RTQTYWDEQRKIYHIQLYLS 212
> sce:YDL043C PRP11, RNA11; Prp11p; K12826 splicing factor 3A
subunit 2
Length=266
Score = 60.1 bits (144), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 59/246 (23%), Positives = 96/246 (39%), Gaps = 67/246 (27%)
Query 19 GSGAPATWQEVNLERKERLRRLALETIDLNKDPYFMRNHLGQFECRLCLTLHTNEGSYLS 78
G P T+Q+ E+ +++R +PY +NH G+ C+LC T+H + S
Sbjct 37 GENVPYTFQD---EKDDQVRS----------NPYIYKNHSGKLVCKLCNTMHMSWSSVER 83
Query 79 HTQGKKHQTNLSRR-LAKEKAD----------------AVVAPMPLKTSSGPGRSTDRVR 121
H GKKH N+ RR ++ EK+ + A LK +
Sbjct 84 HLGGKKHGLNVLRRGISIEKSSLGREGQTTHDFRQQQKIIEAKQSLKNNGTI-------- 135
Query 122 IGRPGYRVTKLRDERTRQFALLCEVEYPEIVRGTK----------PYHRFMSAFEQRVET 171
P ++ +++ + L +V Y V+ P R +S E +T
Sbjct 136 ---PVCKIATVKNPKNGSVGLAIQVNYSSEVKENSVDSDDKAKVPPLIRIVSGLELS-DT 191
Query 172 PPDGNYQFLLFAAEPYETIAFKIPNMEIDRDDP--------------RHHAAWDEEKKVY 217
G +FL+ A EP+E IA ++P EI + + WD K+Y
Sbjct 192 KQKGK-KFLVIAYEPFENIAIELPPNEILFSENNDMDNNNDGVDELNKKCTFWDAISKLY 250
Query 218 TMQIFF 223
+Q FF
Sbjct 251 YVQFFF 256
> tpv:TP04_0385 hypothetical protein; K12848 U4/U6.U5 tri-snRNP
component SNU23
Length=259
Score = 36.6 bits (83), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 34/75 (45%), Gaps = 14/75 (18%)
Query 43 ETIDLNKDPYFMRNHL----------GQFECRLCLTLHTNEGSYLSHTQGKKHQ--TNLS 90
ETID +K Y R+ L G F C+ C L + SYL H GKKH ++
Sbjct 70 ETIDFSK--YVGRSELVDVDGPKSKQGGFFCKTCNCLLKDSKSYLDHLNGKKHNRLLGMT 127
Query 91 RRLAKEKADAVVAPM 105
R+ K A AV +
Sbjct 128 MRVEKVSAKAVADKL 142
> hsa:91603 ZNF830, CCDC16, MGC20398, OMCG1, SEL13; zinc finger
protein 830; K13104 zinc finger protein 830
Length=372
Score = 35.4 bits (80), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 6/63 (9%)
Query 34 KERLRRLALETIDLN------KDPYFMRNHLGQFECRLCLTLHTNEGSYLSHTQGKKHQT 87
+E LRRL E L+ + P+ N LGQ C LC T +E + +H GK+H+
Sbjct 18 QEELRRLMKEKQRLSTSRKRIESPFAKYNRLGQLSCALCNTPVKSELLWQTHVLGKQHRE 77
Query 88 NLS 90
++
Sbjct 78 KVA 80
> ath:AT5G61190 zinc finger protein-related
Length=977
Score = 35.4 bits (80), Expect = 0.22, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 63 CRLCLTLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAP 104
C++C TN +Y +HT GKKH+ NL + K K + +V P
Sbjct 299 CKVCQISFTNNDTYKNHTYGKKHRNNLELQSGKSK-NILVGP 339
Score = 32.7 bits (73), Expect = 1.4, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 31/58 (53%), Gaps = 3/58 (5%)
Query 63 CRLCLTLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRSTDRV 120
C++C ++ ++ SHT GKKH+ NL + AK + ++ P K S G T +V
Sbjct 684 CQVCQISCNSKVAFASHTYGKKHRQNLESQSAKNET---MSTGPGKLSKDYGEKTKKV 738
> mmu:66983 Zfp830, 2410003C20Rik, AV301118, AW048207, Ccdc16,
Omcg1, Znf830; zinc finger protein 830; K13104 zinc finger
protein 830
Length=363
Score = 35.0 bits (79), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 37/74 (50%), Gaps = 9/74 (12%)
Query 23 PATWQEVNLERKERLRRLALETIDLNKD------PYFMRNHLGQFECRLCLTLHTNEGSY 76
PA + VN +E LRRL E L+ + P+ N LGQ C LC T +E +
Sbjct 10 PAGKRVVN---QEELRRLMREKQRLSTNRKRIESPFAKYNRLGQLSCALCNTPVKSELLW 66
Query 77 LSHTQGKKHQTNLS 90
+H GK+H+ ++
Sbjct 67 QTHVLGKQHRERVA 80
> xla:399224 zfr; zinc finger RNA binding protein; K13203 zinc
finger RNA-binding protein
Length=1054
Score = 34.3 bits (77), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 32/71 (45%), Gaps = 1/71 (1%)
Query 17 KLGSGAPATWQEVNLERKERLRRLALE-TIDLNKDPYFMRNHLGQFECRLCLTLHTNEGS 75
K+ P T++E +K + + AL+ + + +R Q C LC T +
Sbjct 328 KISCAGPQTYKEHLEGQKHKKKEAALKASQNTTSSSTAVRGTQNQLRCELCDVSCTGADA 387
Query 76 YLSHTQGKKHQ 86
Y +H +G KHQ
Sbjct 388 YAAHIRGAKHQ 398
> dre:324476 zmat2, fc30d10, flj31121, wu:fc30d10; zinc finger,
matrin type 2; K12848 U4/U6.U5 tri-snRNP component SNU23
Length=198
Score = 34.3 bits (77), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query 58 LGQFECRLCLTLHTNEGSYLSHTQGKKHQTNL--SRRLAKEKADAV 101
+G + C +C + + ++L H GKKHQ NL S R+ + D V
Sbjct 76 MGGYYCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSSLDQV 121
> xla:380471 znf346, DsRBP-ZFa, MGC52555, ZFa, jaz; zinc finger
protein 346
Length=524
Score = 34.3 bits (77), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 61 FECRLCLTLHTNEGSYLSHTQGKKHQTNLSRR--LAKEKADAVVAPMPLKT-SSGPGRST 117
F C C + + Y +H G KH+ L L++E AVVAP + + S+G G S
Sbjct 229 FSCDKCNIVLNSIEQYQAHVSGAKHKNQLMSMTPLSEEGHQAVVAPSAIASGSAGKGFSC 288
Query 118 DRVRI 122
D I
Sbjct 289 DTCNI 293
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 30/65 (46%), Gaps = 3/65 (4%)
Query 61 FECRLCLTLHTNEGSYLSHTQGKKHQTNLSRR--LAKEKADAVVAPMPLKT-SSGPGRST 117
F C C + + Y +H G KH+ +L L++E A VAP + S+G G S
Sbjct 286 FSCDTCNIVLNSIEQYQAHISGAKHKNHLKSMTPLSEEGHTAAVAPSAFASGSAGKGFSC 345
Query 118 DRVRI 122
D I
Sbjct 346 DTCNI 350
> mmu:66492 Zmat2, 2610510D14Rik, 2900082I05Rik; zinc finger,
matrin type 2; K12848 U4/U6.U5 tri-snRNP component SNU23
Length=199
Score = 33.9 bits (76), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 55 RNHLGQFECRLCLTLHTNEGSYLSHTQGKKHQTNL--SRRLAKEKADAV 101
++ +G + C +C + + ++L H GKKHQ NL S R+ + D V
Sbjct 74 QSEMGGYYCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSTLDQV 122
> hsa:153527 ZMAT2, FLJ31121, Snu23, hSNU23; zinc finger, matrin-type
2; K12848 U4/U6.U5 tri-snRNP component SNU23
Length=199
Score = 33.9 bits (76), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 55 RNHLGQFECRLCLTLHTNEGSYLSHTQGKKHQTNL--SRRLAKEKADAV 101
++ +G + C +C + + ++L H GKKHQ NL S R+ + D V
Sbjct 74 QSEMGGYYCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSTLDQV 122
> xla:495824 zmat2, snu23; zinc finger, matrin-type 2; K12848
U4/U6.U5 tri-snRNP component SNU23
Length=199
Score = 33.5 bits (75), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 55 RNHLGQFECRLCLTLHTNEGSYLSHTQGKKHQTNL--SRRLAKEKADAV 101
++ +G + C +C + + ++L H GKKHQ NL S R+ + D V
Sbjct 74 QSDMGGYYCNVCDCVVKDSINFLDHINGKKHQRNLGMSMRVERSTLDQV 122
> ath:AT3G05760 nucleic acid binding / zinc ion binding; K12848
U4/U6.U5 tri-snRNP component SNU23
Length=202
Score = 33.1 bits (74), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 63 CRLCLTLHTNEGSYLSHTQGKKHQTNLSRRLAKEKA 98
CR+C + + +YL H GKKHQ L + E++
Sbjct 85 CRVCDCVVKDSANYLDHINGKKHQRALGMSMRVERS 120
> bbo:BBOV_II002560 18.m06206; hypothetical protein; K12848 U4/U6.U5
tri-snRNP component SNU23
Length=232
Score = 33.1 bits (74), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 35/76 (46%), Gaps = 9/76 (11%)
Query 32 ERKERLRRLALETIDLNKDPYFMR--------NHLGQFECRLCLTLHTNEGSYLSHTQGK 83
E +E L+R E +D+ KD +R + G F C+ C + + +Y+ H G+
Sbjct 62 EVRENLKRRK-EFVDVTKDVGKVRVINALTHKSQQGGFYCKTCDCILKDSQAYMDHLNGR 120
Query 84 KHQTNLSRRLAKEKAD 99
KH L + EK D
Sbjct 121 KHNRMLGMTMRVEKVD 136
> pfa:PFL2075c conserved Plasmodium protein; K12848 U4/U6.U5 tri-snRNP
component SNU23
Length=242
Score = 32.0 bits (71), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 26/92 (28%), Positives = 39/92 (42%), Gaps = 8/92 (8%)
Query 32 ERKERL---RRLALETIDLNKDPYFMRNHLGQFECRLCLTLHTNEGSYLSHTQGKKHQTN 88
ERKE L + L I K P +N G + C++C + + +YL H GK H
Sbjct 83 ERKENLLLEKNLGKVQILTQKTP---KNEQGGYYCKICDCVLKDSQTYLDHINGKNHNRM 139
Query 89 L--SRRLAKEKADAVVAPMPLKTSSGPGRSTD 118
L S ++ K D V + L ++ D
Sbjct 140 LGYSMKVKKVTLDDVKKRLSLLKDKKQNKTED 171
> hsa:23567 ZNF346, DKFZp547M223, JAZ, Zfp346; zinc finger protein
346
Length=294
Score = 31.6 bits (70), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 62 ECRLCLTLHTNEGSYLSHTQGKKHQTNLSRRLA 94
+C++C L +E L+H Q KKH + R LA
Sbjct 74 QCKVCCALLISESQKLAHYQSKKHANKVKRYLA 106
> xla:378688 trove2, ssa2, ssa2-A; TROVE domain family, member
2; K11089 60 kDa SS-A/Ro ribonucleoprotein
Length=538
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 38/90 (42%), Gaps = 10/90 (11%)
Query 31 LERKERLRRLA--LETIDLNKDPYFMRNHLGQFECRLCLTLHTNEGSYLSHTQGKKHQTN 88
LE ER++R LE I L + +R HL LT+H T
Sbjct 229 LEATERVKRTKDELEIIHLIDEYRLVREHL--------LTIHLKSKEIWKSLLQDMPLTA 280
Query 89 LSRRLAKEKADAVVAPMPLKTSSGPGRSTD 118
L R L K AD+V+AP + SS R T+
Sbjct 281 LLRNLGKMTADSVLAPASSEVSSVCERLTN 310
> hsa:51663 ZFR, FLJ41312, ZFR1; zinc finger RNA binding protein;
K13203 zinc finger RNA-binding protein
Length=1074
Score = 31.2 bits (69), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 29/71 (40%), Gaps = 1/71 (1%)
Query 17 KLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFM-RNHLGQFECRLCLTLHTNEGS 75
K+ P T++E +K + + AL+ R Q C LC T +
Sbjct 337 KISCAGPQTYKEHLEGQKHKKKEAALKASQNTSSSNSSTRGTQNQLRCELCDVSCTGADA 396
Query 76 YLSHTQGKKHQ 86
Y +H +G KHQ
Sbjct 397 YAAHIRGAKHQ 407
> mmu:22763 Zfr, C920030H05Rik, MGC176100; zinc finger RNA binding
protein; K13203 zinc finger RNA-binding protein
Length=1074
Score = 31.2 bits (69), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 29/71 (40%), Gaps = 1/71 (1%)
Query 17 KLGSGAPATWQEVNLERKERLRRLALETIDLNKDPYFM-RNHLGQFECRLCLTLHTNEGS 75
K+ P T++E +K + + AL+ R Q C LC T +
Sbjct 337 KISCAGPQTYKEHLEGQKHKKKEAALKASQNTSSSNNSTRGTQNQLRCELCDVSCTGADA 396
Query 76 YLSHTQGKKHQ 86
Y +H +G KHQ
Sbjct 397 YAAHIRGAKHQ 407
> dre:321659 zfr, hnrpa1, wu:fb25b08, zgc:66235, zgc:66403; zinc
finger RNA binding protein; K13203 zinc finger RNA-binding
protein
Length=1052
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 33/75 (44%), Gaps = 6/75 (8%)
Query 17 KLGSGAPATWQEVNLE-----RKERLRRLALETIDLNKDPYFMRNHLGQFECRLCLTLHT 71
K+ P T++E +LE +KE +++ + + R Q C LC T
Sbjct 308 KISCAGPQTYKE-HLEGQKHKKKEAALKVSQSSSSSSGGGSSARGTQNQLRCELCDVSCT 366
Query 72 NEGSYLSHTQGKKHQ 86
+Y +H +G KHQ
Sbjct 367 GADAYAAHIRGAKHQ 381
> mmu:26919 Zfp346, Jaz, Znf346; zinc finger protein 346
Length=294
Score = 30.8 bits (68), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 62 ECRLCLTLHTNEGSYLSHTQGKKHQTNLSRRLA 94
+C++C + +E L+H Q KKH + R LA
Sbjct 74 QCKVCCAMLISESQKLAHYQSKKHANKVKRYLA 106
> cel:Y95B8A.7 hypothetical protein
Length=608
Score = 30.8 bits (68), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 40/88 (45%), Gaps = 9/88 (10%)
Query 12 NRVGHKLGSGAPATWQEVNLER------KERLRRLALETIDLNKDPYFMRNHLGQFECRL 65
+ VG K G A T EV E+ E ++ + E + +D L QF C+L
Sbjct 123 SSVGRKTGGAAATTTIEVEDEKLRAMIAAEEVQPVGEEHVTEERD---ATGKLVQFHCKL 179
Query 66 CLTLHTNEGSYLSHTQGKKHQTNLSRRL 93
C ++ + H +G++H+ + +++
Sbjct 180 CDCKFSDPNAKEIHIKGRRHRVSYRQKI 207
> dre:791686 cadherin 1, type 1 preproprotein-like
Length=777
Score = 30.4 bits (67), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 42/96 (43%), Gaps = 11/96 (11%)
Query 31 LERKERLRRLALETIDLNKDPYFMR-------NHLGQFECRLCLTLHTN--EGSYLS-HT 80
L R ERL R+ T D YF N G R +TLH E S + +
Sbjct 51 LHRGERLGRVRFNTCDRRTRVYFQSTDQEIDLNRDGTLMMRRSVTLHEGFKEFSVTAWDS 110
Query 81 QGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRS 116
GKKH T++ + +D +V P K+S+G R+
Sbjct 111 SGKKHTTSVRVERMESSSDDLVLTFP-KSSTGLKRA 145
> hsa:79842 ZBTB3, FLJ23392; zinc finger and BTB domain containing
3; K10490 zinc finger and BTB domain-containing protein
3
Length=574
Score = 30.0 bits (66), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 5/57 (8%)
Query 61 FECRLCLTLHTNEGSYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRST 117
+ECR CL +T G H + K H +L++R K D V P+ L PG T
Sbjct 500 YECRYCLRSYTQSGDLYRHIR-KAHNEDLAKR---SKPDPEVGPL-LGVQPLPGSPT 551
> dre:567465 zfr2, MGC174987, si:ch211-63j24.3, wu:fb99a09, zfr;
zinc finger RNA binding protein 2
Length=999
Score = 30.0 bits (66), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 26/103 (25%), Positives = 45/103 (43%), Gaps = 8/103 (7%)
Query 17 KLGSGAPATWQEVNLERKERLRRLALETI--DLNKDPYFMRNHLGQFECRLCLTLHTNEG 74
K+ P T++E +LE ++ ++ A + + P R Q C LC T
Sbjct 319 KISCAGPQTYRE-HLEGQKHKKKEAAQKSGSQVTNGP---RGVQTQLRCELCDVSCTGAD 374
Query 75 SYLSHTQGKKHQTNLSRRLAKEKADAVVAPMPLKTSSGPGRST 117
+Y +H +G KHQ + +L + + + P+ SS P T
Sbjct 375 AYAAHIRGSKHQKVV--KLHTKLGKPIPSTEPVLVSSTPAAMT 415
> ath:AT2G19380 RNA recognition motif (RRM)-containing protein
Length=613
Score = 30.0 bits (66), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 60 QFECRLCLTLHTNEGSYLSHTQGKKHQ 86
Q+ C LC T E Y +H GKKHQ
Sbjct 84 QWFCSLCNATMTCEQDYFAHVYGKKHQ 110
Lambda K H
0.321 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8828802264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40