bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3506_orf2
Length=308
Score E
Sequences producing significant alignments: (Bits) Value
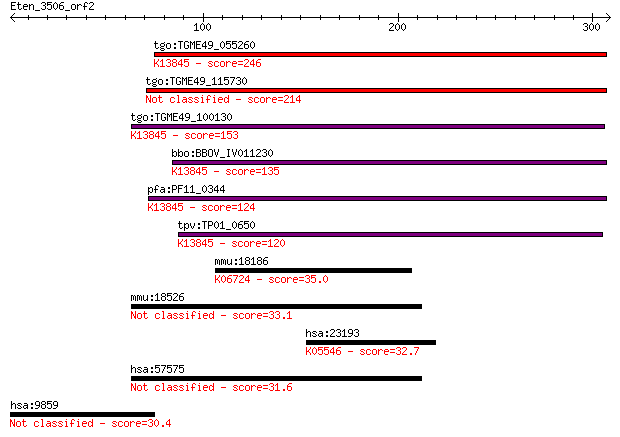
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 246 9e-65
tgo:TGME49_115730 apical membrane antigen, putative 214 4e-55
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 153 8e-37
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 135 2e-31
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 124 3e-28
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 120 7e-27
mmu:18186 Nrp1, C530029I03, NP-1, NPN-1, Npn1, Nrp; neuropilin... 35.0 0.36
mmu:18526 Pcdh10, 6430521D13Rik, 6430703F07Rik, OL-pc, mKIAA14... 33.1 1.2
hsa:23193 GANAB, G2AN, GLUII, KIAA0088; glucosidase, alpha; ne... 32.7 1.8
hsa:57575 PCDH10, DKFZp761O2023, KIAA1400, MGC133344, OL-PCDH,... 31.6 4.4
hsa:9859 CEP170, FAM68A, KAB, KIAA0470; centrosomal protein 17... 30.4 8.0
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 246 bits (627), Expect = 9e-65, Method: Compositional matrix adjust.
Identities = 122/239 (51%), Positives = 158/239 (66%), Gaps = 8/239 (3%)
Query 75 ASTDSNPFM-QPPYAEFMARFNIPKVHGSGVYVDLGNDKEVKGKMYREPGGRCPVFGKNI 133
AST NPF FM RFN+ H SG+YVDLG DKEV G +YREP G CP++GK+I
Sbjct 65 ASTSGNPFQANVEMKTFMERFNLTHHHQSGIYVDLGQDKEVDGTLYREPAGLCPIWGKHI 124
Query 134 EFYQPLDSDLYKNDFLENVPTE-EAAAAAKPLPGGFNNNFLMKDKKPFSPMSVAQLNSYP 192
E QP D Y+N+FLE+VPTE E + PLPGGFN NF+ + SP + L
Sbjct 125 ELQQP-DRPPYRNNFLEDVPTEKEYKQSGNPLPGGFNLNFVTPSGQRISPFPMELLEKNS 183
Query 193 QLKARTGLGKCAEMSYLTTAA-----GSSYRYPFVFDSKKDLCYLLLVPLQRLMGERYCS 247
+KA T LG+CAE ++ T A + YRYPFV+DSKK LC++L V +Q + G++YCS
Sbjct 184 NIKASTDLGRCAEFAFKTVAMDKNNKATKYRYPFVYDSKKRLCHILYVSMQLMEGKKYCS 243
Query 248 TRGSPPGLSHFCFKPLKSVSLRPHLVYGSAYVGERPDDWETKCPNKAVKDAVFGVWEGG 306
+G PP L+ +CFKP KSV+ HL+YGSAYVGE PD + +KCPN+A++ FGVW+ G
Sbjct 244 VKGEPPDLTWYCFKPRKSVTENHHLIYGSAYVGENPDAFISKCPNQALRGYRFGVWKKG 302
> tgo:TGME49_115730 apical membrane antigen, putative
Length=388
Score = 214 bits (544), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 115/245 (46%), Positives = 152/245 (62%), Gaps = 17/245 (6%)
Query 71 ASSDASTDSNPF-MQPPYAEFMARFNIPKVHGSGVYVDLGNDKEVKGKMYREPGGRCPVF 129
A S + NP+ +A+FM RFNIP+VHGSG++VDLG D E YRE GG+CPVF
Sbjct 89 ARSVEAVKQNPWATTTAFADFMKRFNIPQVHGSGIFVDLGRDTE----GYREVGGKCPVF 144
Query 130 GKNIEFYQPLDSDLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKDKKPFSPMSVAQLN 189
GK I+ +QP + Y N+FL++ PT A+ KPLPGGFNN + + FSP+ + L
Sbjct 145 GKAIQMHQPAE---YSNNFLDDAPTSNDASK-KPLPGGFNNPQVYTSGQKFSPIDDSLLQ 200
Query 190 SYPQLKA-RTGLGKCAEMSYLTTAAG------SSYRYPFVFDSKKDLCYLLLVPLQRLMG 242
+T +G+CA +Y T A S+Y+YPFV+D+ CY+L V Q L G
Sbjct 201 ERLGTAGPKTAIGRCALYAYSTIAVNPSTNYTSTYKYPFVYDAVSRKCYVLSVSAQLLKG 260
Query 243 ERYCSTRGSPPGLSHFCFKPLKSVSLRPHLVYGSAYVGE-RPDDWETKCPNKAVKDAVFG 301
E+YCS G+P GL+ CF+P+K S LVYGSA+V E PD W++ CPN AVKDA+FG
Sbjct 261 EKYCSVNGTPSGLTWACFEPVKEKSSARALVYGSAFVAEGNPDAWQSACPNDAVKDALFG 320
Query 302 VWEGG 306
WE G
Sbjct 321 KWEDG 325
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 153 bits (387), Expect = 8e-37, Method: Compositional matrix adjust.
Identities = 97/271 (35%), Positives = 141/271 (52%), Gaps = 43/271 (15%)
Query 63 SHAAAVLAASSDASTDSNPFMQPPYAEFMARFNIPKVHGSGVYVDLGNDKEVKGKMYREP 122
+H + + + A D+NP +A+ + R+N+P VHGSGVYVDLGN K + K YREP
Sbjct 38 AHQTRLASGKTSAKGDANP-----WAKILERYNVPLVHGSGVYVDLGNTKILSKKKYREP 92
Query 123 GGRCPVFGKNIEFYQPLDS-DLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKDKKPFS 181
GG+CP +GK I+ YQP + +++ NDFL+ VP PL GGF S
Sbjct 93 GGKCPNYGKYIKTYQPTTNPEIWPNDFLKPVPYANTPQDTMPLGGGF-----AMPMHQIS 147
Query 182 PMSVAQLNSYPQ-LKARTG---------------LGKCAEMSYLTTAAGSS--------- 216
P+S+ L + LK TG LG C + +T+A +S
Sbjct 148 PVSLKDLKDEAEGLKTATGVSSYAVEHAKNIRDDLGHCIWWARMTSAHDTSSSATNSKED 207
Query 217 -YRYPFVFDSKKDLCYLLLVPLQRLMGE-RYCSTRGSPPGLSHFCFKPLKSVSLRPHLVY 274
YRY FV+D KK++C+++ + +Q + G YC S P L+ +CF P KS+ +LV+
Sbjct 208 YYRYAFVWDPKKEMCHIMYLNMQEMTGAGTYCKRGDSGPNLTWYCFHPEKSI--EKNLVW 265
Query 275 GSAYVGERPDDWETKCPNKAVKDAVFGVWEG 305
GSAY R D + CP +K+ +G W G
Sbjct 266 GSAYA--RLDH-ASACPEHGLKNVHWGQWNG 293
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 135 bits (340), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 88/236 (37%), Positives = 120/236 (50%), Gaps = 24/236 (10%)
Query 84 QPPYAEFMARFNIPKVHGSGVYVDLGNDKEVKGKMYREPGGRCPVFGKNIEFYQPLDSDL 143
Q P+ ++M +F+IP+ HGSG+YVDLG + V K YR P G+CPV GK I+
Sbjct 94 QSPWIKYMQKFDIPRNHGSGIYVDLGGYESVGSKSYRMPVGKCPVVGKIIDLGNGA---- 149
Query 144 YKNDFLENVPTEEAAAAAKPLP-GGFNNNFLMKDK-KPFSPMSVAQLNSYPQLKAR---- 197
DFL+ + +E+ + P ++N + K + S ++ A+L+ R
Sbjct 150 ---DFLDPISSEDPSYRGLAFPETAVDSNIPTQPKTRGSSSVTAAKLSPVSAKDLRRWGY 206
Query 198 --TGLGKCAE-MSYLTTAAG--SSYRYPFVFDSKKDLCYLLLVPLQRLMGERYCSTRGSP 252
+ C+E S L A+ + YRYPFVFDS +CY+L +Q G RYC GS
Sbjct 207 EGNDVANCSEYASNLIPASDKTTKYRYPFVFDSDNQMCYILYSAIQYNQGNRYCDNDGSS 266
Query 253 P--GLSHFCFKPLKSVSLRPHLVYGSAYVGERPDDWETKCPNKAVKDAVFGVWEGG 306
S C KP KS HL YGSA V DWE CP V+DA+FG W GG
Sbjct 267 EEGTSSLLCMKPYKSAE-DAHLYYGSAKVDP---DWEENCPMHPVRDAIFGKWSGG 318
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 124 bits (312), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 79/245 (32%), Positives = 118/245 (48%), Gaps = 31/245 (12%)
Query 72 SSDASTDSNPFMQPPYAEFMARFNIPKVHGSGVYVDLGNDKEVKGKMYREPGGRCPVFGK 131
SS + + +M P+ E+MA+++I +VHGSG+ VDLG D EV G YR P G+CPVFGK
Sbjct 95 SSIEIVERSNYMGNPWTEYMAKYDIEEVHGSGIRVDLGEDAEVAGTQYRLPSGKCPVFGK 154
Query 132 NIEFYQPLDSDLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKDKKPF-SPMSVAQLNS 190
I +EN T A + F +P SPM++ ++
Sbjct 155 GI--------------IIENSNTTFLTPVATGNQYLKDGGFAFPPTEPLMSPMTLDEMRH 200
Query 191 YPQLKARTGLGKCAEMSYLTTAAG---------SSYRYPFVFDSKKDLCYLLLVPLQRLM 241
+ K + E++ + AG S+Y+YP V+D K C++L + Q
Sbjct 201 F--YKDNKYVKNLDELTLCSRHAGNMIPDNDKNSNYKYPAVYDDKDKKCHILYIAAQENN 258
Query 242 GERYCSTRGSPPGLSHFCFKPLKSVSLRPHLVYGSAYVGERPDDWETKCPNKAVKDAVFG 301
G RYC+ S S FCF+P K +S + + V D+WE CP K +++A FG
Sbjct 259 GPRYCNKDESKRN-SMFCFRPAKDISFQNYTYLSKNVV----DNWEKVCPRKNLQNAKFG 313
Query 302 VWEGG 306
+W G
Sbjct 314 LWVDG 318
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 120 bits (300), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 80/227 (35%), Positives = 112/227 (49%), Gaps = 23/227 (10%)
Query 87 YAEFMARFNIPKVHGSGVYVDLGNDKEVKGKMYREPGGRCPVFGKNIEFYQPLDSDLYKN 146
+ EFMA+F+I KVHGSGVYVDLG V YR P G+CPV GK I + +N
Sbjct 250 WTEFMAKFDIAKVHGSGVYVDLGESATVGIYDYRMPIGKCPVVGKAI---------ILEN 300
Query 147 --DFLENVPTEEAAAAAKPLPG---GFNNNFLMKDKKPFSPMSVAQLNSYPQLKARTGLG 201
DFL ++ + P N++ + + SP+S L S+ K + L
Sbjct 301 GADFLSSITHHDPKERGLGFPATKVASNSSKQDMENQLLSPISAQVLRSW-NYKHESDLS 359
Query 202 KCAEMSYLT---TAAGSSYRYPFVFDSKKDLCYLLLVPLQRLMGERYCSTRGSPPGLSHF 258
CAE S + S YRYPFV+D + LCY+L P+Q G +YC + G S
Sbjct 360 NCAEYSRNIVPGSNRNSKYRYPFVYDESEKLCYILYSPMQYNQGVKYCDKDSADEGTSSL 419
Query 259 -CFKPLKSVSLRPHLVYGSAYVGERPDDWETKCPNKAVKDAVFGVWE 304
C P KS HL YG++ + DW CP ++D++FG ++
Sbjct 420 ACMYPDKSKD-DSHLFYGTSGLHM---DWPVVCPVYPIRDSIFGSYD 462
> mmu:18186 Nrp1, C530029I03, NP-1, NPN-1, Npn1, Nrp; neuropilin
1; K06724 neuropilin 1
Length=923
Score = 35.0 bits (79), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 47/101 (46%), Gaps = 7/101 (6%)
Query 106 VDLGNDKEVKGKMYREPGGRCPVFGKNIEFYQPLDSDLYKNDFLENVPTEEAAAAAKPLP 165
VDLG++K V+G + + GG+ +N F + N +++ AK
Sbjct 487 VDLGDEKIVRGVIIQ--GGK---HRENKVFMRKFKIAYSNNGSDWKTIMDDSKRKAKSFE 541
Query 166 GGFNNNFLMKDKKPFSPMSVAQLNSYPQLKARTGLGKCAEM 206
G NNN+ + + FSP+S + YP+ +GLG E+
Sbjct 542 G--NNNYDTPELRTFSPLSTRFIRIYPERATHSGLGLRMEL 580
> mmu:18526 Pcdh10, 6430521D13Rik, 6430703F07Rik, OL-pc, mKIAA1400;
protocadherin 10
Length=1057
Score = 33.1 bits (74), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 41/162 (25%), Positives = 65/162 (40%), Gaps = 17/162 (10%)
Query 63 SHAAAVLAASSDASTDSNPFMQPPYAEFMARFNIPKVHGSGVYVDLGNDK-EVKGKMYRE 121
S + ++ SD + ++ F QP Y ++ N+P G+ +Y D+ E
Sbjct 444 STSKSIQVQVSDVNDNAPRFSQPVYDVYVTENNVP---GAYIYAVSATDRDEGANAKLTY 500
Query 122 PGGRCPVFGKNIEFYQPLDSD---LYK---------NDFLENVPTEEAAAAAKPLPGGFN 169
C + G ++ Y ++SD LY DF V + A + + L G
Sbjct 501 SILECQIQGMSVFTYVSINSDNGYLYALRSFDYEQIKDFSFQVEARD-AGSPQALAGNAT 559
Query 170 NNFLMKDKKPFSPMSVAQLNSYPQLKARTGLGKCAEMSYLTT 211
N L+ D+ +P VA L AR L + AE YL T
Sbjct 560 VNILIVDQNDNAPAIVAPLPGRNGTPAREVLPRSAEPGYLLT 601
> hsa:23193 GANAB, G2AN, GLUII, KIAA0088; glucosidase, alpha;
neutral AB (EC:3.2.1.84); K05546 alpha 1,3-glucosidase [EC:3.2.1.84]
Length=944
Score = 32.7 bits (73), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 30/68 (44%), Gaps = 2/68 (2%)
Query 153 PTEEAAAAAKPLPGGFNNNF-LMKDKKPFSPMSVAQLNSYPQLKARTGLGKCAEMSYL-T 210
P E A K PG + F D KP+ PMSV S P ++ G+ + A+ L
Sbjct 211 PEETQGKAEKDEPGAWEETFKTHSDSKPYGPMSVGLDFSLPGMEHVYGIPEHADNLRLKV 270
Query 211 TAAGSSYR 218
T G YR
Sbjct 271 TEGGEPYR 278
> hsa:57575 PCDH10, DKFZp761O2023, KIAA1400, MGC133344, OL-PCDH,
PCDH19; protocadherin 10
Length=896
Score = 31.6 bits (70), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 40/162 (24%), Positives = 65/162 (40%), Gaps = 17/162 (10%)
Query 63 SHAAAVLAASSDASTDSNPFMQPPYAEFMARFNIPKVHGSGVYVDLGNDK-EVKGKMYRE 121
S + ++ SD + ++ F QP Y ++ N+P G+ +Y D+ E
Sbjct 444 STSKSIQVQVSDVNDNAPRFSQPVYDVYVTENNVP---GAYIYAVSATDRDEGANAQLAY 500
Query 122 PGGRCPVFGKNIEFYQPLDSD---LYK---------NDFLENVPTEEAAAAAKPLPGGFN 169
C + G ++ Y ++S+ LY DF V + A + + L G
Sbjct 501 SILECQIQGMSVFTYVSINSENGYLYALRSFDYEQLKDFSFQVEARD-AGSPQALAGNAT 559
Query 170 NNFLMKDKKPFSPMSVAQLNSYPQLKARTGLGKCAEMSYLTT 211
N L+ D+ +P VA L AR L + AE YL T
Sbjct 560 VNILIVDQNDNAPAIVAPLPGRNGTPAREVLPRSAEPGYLLT 601
> hsa:9859 CEP170, FAM68A, KAB, KIAA0470; centrosomal protein
170kDa
Length=1486
Score = 30.4 bits (67), Expect = 8.0, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 30/74 (40%), Gaps = 2/74 (2%)
Query 1 FYSKFEKKKSQNLKRPPSTMRRLSPALGLLAAALSCAGPAAGVQHKLQHRQQQQQQHSHA 60
+ S E + N P T R SPAL L AG A +HR ++Q+ +
Sbjct 1139 YASTSEDEFGSNRNSPKHTRLRTSPALK--TTRLQSAGSAMPTSSSFKHRIKEQEDYIRD 1196
Query 61 STSHAAAVLAASSD 74
T+H + S D
Sbjct 1197 WTAHREEIARISQD 1210
Lambda K H
0.318 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12474975856
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40