bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3480_orf2
Length=159
Score E
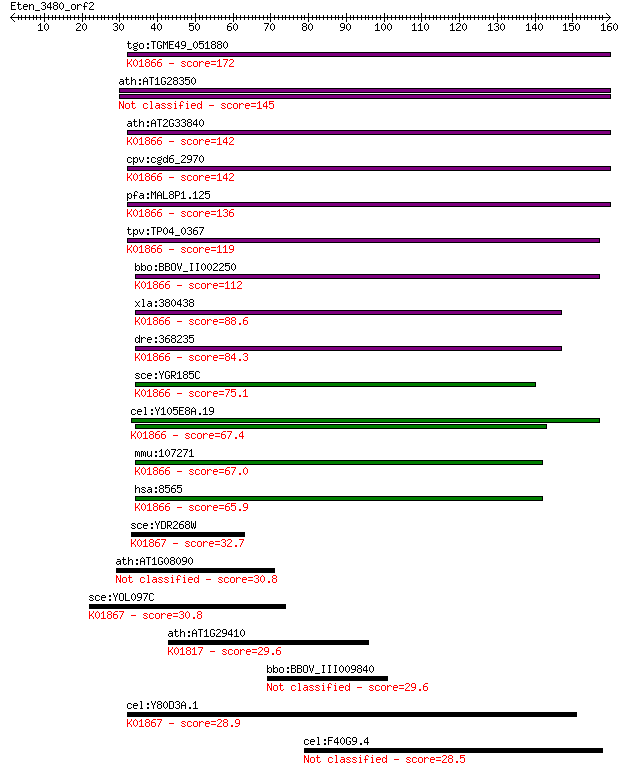
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_051880 tyrosyl-tRNA synthetase, putative (EC:6.1.1.... 172 3e-43
ath:AT1G28350 ATP binding / aminoacyl-tRNA ligase/ nucleotide ... 145 7e-35
ath:AT2G33840 tRNA synthetase class I (W and Y) family protein... 142 3e-34
cpv:cgd6_2970 tyrosyl-tRNA synthetase (tyrosyl-tRNA ligase; Ty... 142 5e-34
pfa:MAL8P1.125 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);... 136 3e-32
tpv:TP04_0367 tyrosyl-tRNA synthetase (EC:6.1.1.1); K01866 tyr... 119 4e-27
bbo:BBOV_II002250 18.m06182; tyrosyl-tRNA synthetase; K01866 t... 112 7e-25
xla:380438 yars, MGC53284; tyrosyl-tRNA synthetase (EC:6.1.1.1... 88.6 8e-18
dre:368235 yars, cb204, wu:fk52b08; tyrosyl-tRNA synthetase (E... 84.3 2e-16
sce:YGR185C TYS1, TTS1; Cytoplasmic tyrosyl-tRNA synthetase, r... 75.1 9e-14
cel:Y105E8A.19 hypothetical protein; K01866 tyrosyl-tRNA synth... 67.4 2e-11
mmu:107271 Yars, AL024047, AU018965; tyrosyl-tRNA synthetase (... 67.0 3e-11
hsa:8565 YARS, CMTDIC, TYRRS, YRS, YTS; tyrosyl-tRNA synthetas... 65.9 6e-11
sce:YDR268W MSW1; Msw1p (EC:6.1.1.2); K01867 tryptophanyl-tRNA... 32.7 0.53
ath:AT1G08090 ATNRT2:1 (NITRATE TRANSPORTER 2:1); nitrate tran... 30.8 1.9
sce:YOL097C WRS1, HRE342; Wrs1p (EC:6.1.1.2); K01867 tryptopha... 30.8 1.9
ath:AT1G29410 PAI3; PAI3 (phosphoribosylanthranilate isomerase... 29.6 4.1
bbo:BBOV_III009840 17.m07852; hypothetical protein 29.6 4.3
cel:Y80D3A.1 wrs-1; tryptophanyl (W) tRNA Synthetase family me... 28.9 7.2
cel:F40G9.4 btb-1; BTB (Broad/complex/Tramtrack/Bric a brac) d... 28.5 8.9
> tgo:TGME49_051880 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=427
Score = 172 bits (437), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 78/129 (60%), Positives = 103/129 (79%), Gaps = 1/129 (0%)
Query 32 RKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPD-KEFHVQR 90
++KMSKSN DSAIFMED+E EVNRKI+KAFCP + GNPCV YVE+LVFP EF V+R
Sbjct 299 QEKMSKSNPDSAIFMEDTETEVNRKIKKAFCPAGVIDGNPCVTYVEQLVFPAFGEFAVKR 358
Query 91 TLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKLLQ 150
+ NGG++ + T EEF+ ++ G LHP DLK +LARE+NR+LEP+R+HF +E+AK+LLQ
Sbjct 359 SEANGGDVTYATVEEFRNAYIKGNLHPGDLKESLARELNRRLEPIRKHFTENEHAKQLLQ 418
Query 151 EISKYKITR 159
+I YK+T+
Sbjct 419 QIKAYKVTK 427
> ath:AT1G28350 ATP binding / aminoacyl-tRNA ligase/ nucleotide
binding / tyrosine-tRNA ligase
Length=776
Score = 145 bits (365), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 68/131 (51%), Positives = 95/131 (72%), Gaps = 1/131 (0%)
Query 30 KDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP-DKEFHV 88
+ +KKMSKS+ SAIFMED EAEVN KI+KA+CPP V GNPC+ YV+ ++ P EF V
Sbjct 269 QGQKKMSKSDPSSAIFMEDEEAEVNVKIKKAYCPPDIVEGNPCLEYVKHIILPWFSEFTV 328
Query 89 QRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKL 148
+R + GGN FK+ E+ + +G+LHP DLK AL++ +N+ L+PVR HF+T+ AK L
Sbjct 329 ERDEKYGGNRTFKSFEDITTDYESGQLHPKDLKDALSKALNKILQPVRDHFKTNSRAKNL 388
Query 149 LQEISKYKITR 159
L+++ YK+TR
Sbjct 389 LKQVKGYKVTR 399
Score = 137 bits (345), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 66/131 (50%), Positives = 91/131 (69%), Gaps = 2/131 (1%)
Query 30 KDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPD-KEFHV 88
+ ++KMSKS+ SAIFMED EA+VN KI KA+CPP V GNPC+ YV+ +V P EF V
Sbjct 647 QGQEKMSKSDPSSAIFMEDEEADVNEKISKAYCPPKTVEGNPCLEYVKYIVLPRFNEFKV 706
Query 89 QRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKL 148
+ +NGGN F + E+ + GELHP DLK AL + +N L+PVR HF+T+E AK L
Sbjct 707 ESE-KNGGNKTFNSFEDIVADYETGELHPEDLKKALMKALNITLQPVRDHFKTNERAKNL 765
Query 149 LQEISKYKITR 159
L+++ +++TR
Sbjct 766 LEQVKAFRVTR 776
> ath:AT2G33840 tRNA synthetase class I (W and Y) family protein;
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=385
Score = 142 bits (359), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 67/129 (51%), Positives = 91/129 (70%), Gaps = 1/129 (0%)
Query 32 RKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP-DKEFHVQR 90
++KMSKS+ SAIFMED EAEVN KI+KA+CPP V GNPC+ Y++ ++ P EF V+R
Sbjct 257 QEKMSKSDPLSAIFMEDEEAEVNVKIKKAYCPPKVVKGNPCLEYIKYIILPWFDEFTVER 316
Query 91 TLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKLLQ 150
E GGN +K+ E+ + +GELHP DLK L +N+ L+PVR HF+TD AK LL+
Sbjct 317 NEEYGGNKTYKSFEDIAADYESGELHPGDLKKGLMNALNKILQPVRDHFKTDARAKNLLK 376
Query 151 EISKYKITR 159
+I Y++TR
Sbjct 377 QIKAYRVTR 385
> cpv:cgd6_2970 tyrosyl-tRNA synthetase (tyrosyl-tRNA ligase;
TyrRS). class-I aaRS ; K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=376
Score = 142 bits (358), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 63/129 (48%), Positives = 89/129 (68%), Gaps = 1/129 (0%)
Query 32 RKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPD-KEFHVQR 90
++KMSKS+ SAIF+ED+ V +KI+KAFCPP + GNPC+ Y+ LVFP FHV R
Sbjct 248 QEKMSKSDTSSAIFVEDTPEAVVKKIKKAFCPPGIIEGNPCIEYINTLVFPKFGHFHVSR 307
Query 91 TLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKLLQ 150
E GG+I F E+F +++ +G+LHP DLK L+ +N L+P+R HF TD AK+LLQ
Sbjct 308 KEEYGGDITFTNKEDFHKAYLSGDLHPGDLKKGLSDALNLMLQPIRDHFNTDPRAKELLQ 367
Query 151 EISKYKITR 159
+ +K+T+
Sbjct 368 LVQSFKVTK 376
> pfa:MAL8P1.125 tyrosyl-tRNA synthetase, putative (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=373
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 63/129 (48%), Positives = 91/129 (70%), Gaps = 1/129 (0%)
Query 32 RKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPD-KEFHVQR 90
++KMSKS+ +SAIFM+DSE++VNRKI+KA+CPP + NP AY + ++FP EF++ R
Sbjct 245 QEKMSKSDENSAIFMDDSESDVNRKIKKAYCPPNVIENNPIYAYAKSIIFPSYNEFNLVR 304
Query 91 TLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKLLQ 150
+NGG+ + T +E + + G +HP DLK +A IN+ L+PVR HFQ + AK LL
Sbjct 305 KEKNGGDKTYYTLQELEHDYVNGFIHPLDLKDNVAMYINKLLQPVRDHFQNNIEAKNLLN 364
Query 151 EISKYKITR 159
EI KYK+T+
Sbjct 365 EIKKYKVTK 373
> tpv:TP04_0367 tyrosyl-tRNA synthetase (EC:6.1.1.1); K01866 tyrosyl-tRNA
synthetase [EC:6.1.1.1]
Length=427
Score = 119 bits (299), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 57/128 (44%), Positives = 89/128 (69%), Gaps = 5/128 (3%)
Query 32 RKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKEFH---V 88
++KMSKSN +SAIFM+DS EV+ KI+KA+CPP V GNP +AY +VF K F+ +
Sbjct 297 QEKMSKSNPNSAIFMDDSPEEVSSKIKKAYCPPGVVDGNPVIAYFCFIVF--KRFNSVTI 354
Query 89 QRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAKKL 148
+R ++GG++ F + EE K SF G LHPADLK +L + +N ++P+R +F+ ++ AK L
Sbjct 355 ERKEKDGGDVTFNSPEELKESFLQGLLHPADLKNSLVKYLNLMMQPIRDYFEHNQEAKVL 414
Query 149 LQEISKYK 156
++ +++
Sbjct 415 EMKVKQFQ 422
> bbo:BBOV_II002250 18.m06182; tyrosyl-tRNA synthetase; K01866
tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=418
Score = 112 bits (279), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 59/135 (43%), Positives = 84/135 (62%), Gaps = 12/135 (8%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPD-KEFHVQRTL 92
KMSKS +SAIFMEDS EVN KI+ A+CP +V NPC+AY E +VFP + R
Sbjct 281 KMSKSIPNSAIFMEDSAEEVNAKIKSAWCPEKEVCDNPCIAYFEHIVFPMFNTVTIPRKE 340
Query 93 ENG-----------GNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQT 141
+NG G+++++ E K+ + +G LHPADLK ALA INR LEPVR +F+
Sbjct 341 KNGGKRDTSTSKNSGDVIYQCHSELKQDYLSGNLHPADLKPALASYINRLLEPVRIYFRD 400
Query 142 DENAKKLLQEISKYK 156
+ A +L + +++ +
Sbjct 401 NPEANELARRVAELQ 415
> xla:380438 yars, MGC53284; tyrosyl-tRNA synthetase (EC:6.1.1.1);
K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=528
Score = 88.6 bits (218), Expect = 8e-18, Method: Composition-based stats.
Identities = 45/114 (39%), Positives = 68/114 (59%), Gaps = 1/114 (0%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDK-EFHVQRTL 92
KMS S +S I + DS A+V +K++KAFC P V N +++V ++FP K EF V R
Sbjct 222 KMSSSEEESKIDLLDSPADVKKKLKKAFCEPGNVENNGVLSFVRHVLFPLKSEFVVLRDE 281
Query 93 ENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAK 146
+ GGN + E ++ FA +HP DLKA++ + +N+ L P+R F + E K
Sbjct 282 KFGGNKTYTDFETLEKDFAEELVHPGDLKASVEKALNKLLHPIREKFNSPEMKK 335
> dre:368235 yars, cb204, wu:fk52b08; tyrosyl-tRNA synthetase
(EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=529
Score = 84.3 bits (207), Expect = 2e-16, Method: Composition-based stats.
Identities = 43/114 (37%), Positives = 73/114 (64%), Gaps = 1/114 (0%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP-DKEFHVQRTL 92
KMS S +S I + D +V +K++KAFC P V N +++V+ ++FP EF ++R
Sbjct 222 KMSSSEEESKIDLLDKNQDVKKKLKKAFCEPGNVENNGVLSFVKHVLFPLHSEFVIKRDP 281
Query 93 ENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTDENAK 146
+ GG+ V+ EE ++ FAA ++HP DLKA++ +N+ L+P+R+ F++ E K
Sbjct 282 KFGGDKVYTDFEEVEKDFAAEQIHPGDLKASVELALNKLLDPIRKKFESPELKK 335
> sce:YGR185C TYS1, TTS1; Cytoplasmic tyrosyl-tRNA synthetase,
required for cytoplasmic protein synthesis; interacts with
positions 34 and 35 of the tRNATyr anticodon; mutations in human
ortholog YARS are associated with Charcot-Marie-Tooth (CMT)
neuropathies (EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase
[EC:6.1.1.1]
Length=394
Score = 75.1 bits (183), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 63/115 (54%), Gaps = 9/115 (7%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKE-------- 85
KMS S+ +S I + + +V +KI AFC P V N +++V+ ++ P +E
Sbjct 227 KMSASDPNSKIDLLEEPKQVKKKINSAFCSPGNVEENGLLSFVQYVIAPIQELKFGTNHF 286
Query 86 -FHVQRTLENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHF 139
F + R + GG I +K+ EE K +F +L P DLK +A IN LEP+R+ F
Sbjct 287 EFFIDRPEKFGGPITYKSFEEMKLAFKEEKLSPPDLKIGVADAINELLEPIRQEF 341
> cel:Y105E8A.19 hypothetical protein; K01866 tyrosyl-tRNA synthetase
[EC:6.1.1.1]
Length=722
Score = 67.4 bits (163), Expect = 2e-11, Method: Composition-based stats.
Identities = 42/134 (31%), Positives = 63/134 (47%), Gaps = 10/134 (7%)
Query 33 KKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFP---DKEFHVQ 89
+KMS S D + D+ ++ KI ++FC P + GN + E +VFP H+Q
Sbjct 585 QKMSCSIPDFLLDPLDTPKQLKTKIARSFCEPQNLDGNVAMQLAEHIVFPILNGAALHIQ 644
Query 90 RTLENGGNIVFKTAEEFKRSFAAGE-------LHPADLKAALAREINRQLEPVRRHFQTD 142
R +NGG++ ++ ++ F G LHPADLK A+ INR + VR F
Sbjct 645 RAPDNGGDVEVTNYKQLEQEFVIGTKNEQQFPLHPADLKNAVVGVINRLFDGVRADFAEK 704
Query 143 ENAKKLLQEISKYK 156
K + S K
Sbjct 705 PRQKLVTDAFSTSK 718
Score = 33.9 bits (76), Expect = 0.22, Method: Composition-based stats.
Identities = 34/109 (31%), Positives = 46/109 (42%), Gaps = 5/109 (4%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKEFHVQRTLE 93
KMS S DS I + D +V KI A C Q N +++ ++FP V
Sbjct 233 KMSSSEEDSKIDVLDEPEKVRSKIMGAACSRDQ-PDNGVLSFYNFVLFPI----VSPNAI 287
Query 94 NGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQTD 142
N F + KR++ G+L LK LA + LE VR TD
Sbjct 288 EIENQQFFDIDSLKRAYLDGKLDENTLKTFLADFLVNLLEKVRVRCDTD 336
> mmu:107271 Yars, AL024047, AU018965; tyrosyl-tRNA synthetase
(EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=528
Score = 67.0 bits (162), Expect = 3e-11, Method: Composition-based stats.
Identities = 40/109 (36%), Positives = 66/109 (60%), Gaps = 1/109 (0%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDK-EFHVQRTL 92
KMS S +S I + D + +V +K++KAFC P V N +++++ ++FP K EF + R
Sbjct 222 KMSSSEEESKIDLLDRKEDVKKKLKKAFCEPGNVENNGVLSFIKHVLFPLKSEFVILRDE 281
Query 93 ENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQT 141
+ GGN + E ++ FAA +HP DLK ++ +N+ L+P+R F T
Sbjct 282 KWGGNKTYTVYLELEKDFAAEVVHPGDLKNSVEVALNKLLDPIREKFNT 330
> hsa:8565 YARS, CMTDIC, TYRRS, YRS, YTS; tyrosyl-tRNA synthetase
(EC:6.1.1.1); K01866 tyrosyl-tRNA synthetase [EC:6.1.1.1]
Length=528
Score = 65.9 bits (159), Expect = 6e-11, Method: Composition-based stats.
Identities = 39/109 (35%), Positives = 66/109 (60%), Gaps = 1/109 (0%)
Query 34 KMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDK-EFHVQRTL 92
KMS S +S I + D + +V +K++KAFC P V N +++++ ++FP K EF + R
Sbjct 222 KMSSSEEESKIDLLDRKEDVKKKLKKAFCEPGNVENNGVLSFIKHVLFPLKSEFVILRDE 281
Query 93 ENGGNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQLEPVRRHFQT 141
+ GGN + + ++ FAA +HP DLK ++ +N+ L+P+R F T
Sbjct 282 KWGGNKTYTAYVDLEKDFAAEVVHPGDLKNSVEVALNKLLDPIREKFNT 330
> sce:YDR268W MSW1; Msw1p (EC:6.1.1.2); K01867 tryptophanyl-tRNA
synthetase [EC:6.1.1.2]
Length=379
Score = 32.7 bits (73), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 20/32 (62%), Gaps = 2/32 (6%)
Query 33 KKMSKS--NADSAIFMEDSEAEVNRKIRKAFC 62
KKMSKS N DS IF+ D + +KIRKA
Sbjct 243 KKMSKSDPNHDSVIFLNDEPKAIQKKIRKALT 274
> ath:AT1G08090 ATNRT2:1 (NITRATE TRANSPORTER 2:1); nitrate transmembrane
transporter
Length=530
Score = 30.8 bits (68), Expect = 1.9, Method: Composition-based stats.
Identities = 12/42 (28%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 29 WKDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGN 70
W +++K + S F E++++E R++R A PP N
Sbjct 488 WNEQEKQKNMHQGSLRFAENAKSEGGRRVRSAATPPENTPNN 529
> sce:YOL097C WRS1, HRE342; Wrs1p (EC:6.1.1.2); K01867 tryptophanyl-tRNA
synthetase [EC:6.1.1.2]
Length=432
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 27/60 (45%), Gaps = 8/60 (13%)
Query 22 TRCFRDCWKDRKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVH--------GNPCV 73
+R F KMS S+ +AIFM D+ ++ +KI K QV GNP V
Sbjct 283 SRFFPALQGSTTKMSASDDTTAIFMTDTPKQIQKKINKYAFSGGQVSADLHRELGGNPDV 342
> ath:AT1G29410 PAI3; PAI3 (phosphoribosylanthranilate isomerase
3); phosphoribosylanthranilate isomerase (EC:5.3.1.24); K01817
phosphoribosylanthranilate isomerase [EC:5.3.1.24]
Length=244
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 24/53 (45%), Gaps = 0/53 (0%)
Query 43 AIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKEFHVQRTLENG 95
+F+ED E + R + Q+HGN A RLV K +V E+G
Sbjct 123 GVFVEDDENTILRAADSSDLELVQLHGNSSRAAFSRLVRERKVIYVLNANEDG 175
> bbo:BBOV_III009840 17.m07852; hypothetical protein
Length=212
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Query 69 GNPCVAYVERLVFPDKEFHVQRT---LENGGNIVF 100
GN CV Y RL+F D++ +V + +E GGN+ F
Sbjct 160 GNTCVQYRGRLMFFDRDSNVILSDVKVEEGGNLPF 194
> cel:Y80D3A.1 wrs-1; tryptophanyl (W) tRNA Synthetase family
member (wrs-1); K01867 tryptophanyl-tRNA synthetase [EC:6.1.1.2]
Length=417
Score = 28.9 bits (63), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 30/135 (22%), Positives = 51/135 (37%), Gaps = 35/135 (25%)
Query 32 RKKMSKSNADSAIFMEDSEAEVNRKIRKAFCPPAQVHGNPCVAYVERLVFPDKEFHVQRT 91
+ KMS S ++ IF+ D+ ++ KI K F + VQ
Sbjct 294 QTKMSASEPNTCIFLSDTAKQIKNKINK-------------------YAFSGGQQTVQEH 334
Query 92 LENGGNIVFKTAEEFKRSF--------------AAGELHPADLKAALAREINRQLEPV-- 135
E GGN + +F R F GE+ +LKA +++ + +
Sbjct 335 REKGGNCDVDISYQFLRFFLDDDEKLAEIRENYTKGEMLSGELKALATQKVQEIVLEMQE 394
Query 136 RRHFQTDENAKKLLQ 150
RR TDE ++ ++
Sbjct 395 RRKLVTDETVEEFVK 409
> cel:F40G9.4 btb-1; BTB (Broad/complex/Tramtrack/Bric a brac)
domain protein family member (btb-1)
Length=229
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 39/85 (45%), Gaps = 12/85 (14%)
Query 79 LVFPDKEFHVQRTLENG------GNIVFKTAEEFKRSFAAGELHPADLKAALAREINRQL 132
L+ D + HV + + G F++ E+ K F ++ D+KA LA +
Sbjct 72 LLVEDTKLHVNKAFLSYHSEFFRGLFEFESEEKQKNEFKISDVSADDMKAFLAI-----I 126
Query 133 EPVRRHFQTDENAKKLLQEISKYKI 157
P F TD NAKKLL+ ++ +
Sbjct 127 HP-GNDFPTDSNAKKLLELADRFMV 150
Lambda K H
0.320 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40