bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
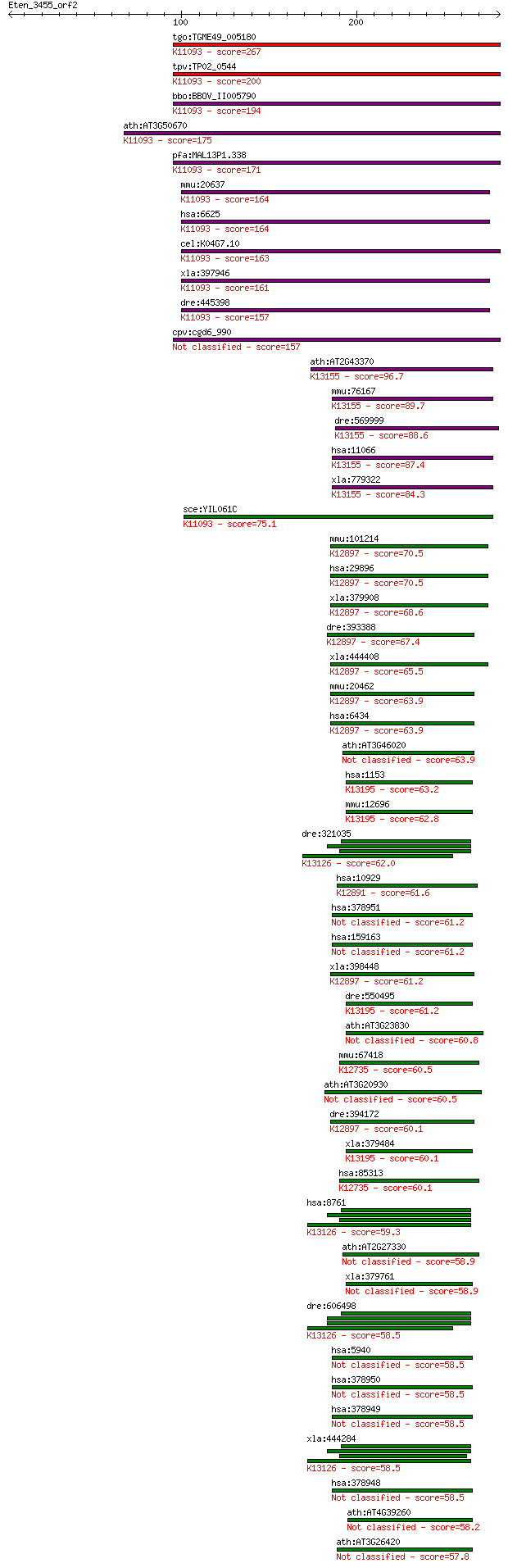
Query= Eten_3455_orf2
Length=281
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_005180 U1 small nuclear ribonucleoprotein, putative... 267 3e-71
tpv:TP02_0544 U1 small nuclear ribonucleoprotein; K11093 U1 sm... 200 4e-51
bbo:BBOV_II005790 18.m06481; u1 snRNP; K11093 U1 small nuclear... 194 3e-49
ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTE... 175 1e-43
pfa:MAL13P1.338 U1 small nuclear ribonucleoprotein, putative; ... 171 4e-42
mmu:20637 Snrnp70, 2700022N21Rik, 3200002N22Rik, 70kDa, AI3250... 164 3e-40
hsa:6625 SNRNP70, RNPU1Z, RPU1, SNRP70, Snp1, U1-70K, U170K, U... 164 3e-40
cel:K04G7.10 rnp-7; RNP (RRM RNA binding domain) containing fa... 163 7e-40
xla:397946 snrnp70, MGC130688, snrp70; small nuclear ribonucle... 161 2e-39
dre:445398 snrnp70, MGC136450, snrp70, wu:fc20b04, zgc:136450;... 157 4e-38
cpv:cgd6_990 U1 snrp Snp1p. RRM domain containing protein 157 6e-38
ath:AT2G43370 U1 small nuclear ribonucleoprotein 70 kDa, putat... 96.7 1e-19
mmu:76167 Snrnp35, 6330548G22Rik; small nuclear ribonucleoprot... 89.7 1e-17
dre:569999 snrnp35, MGC112337, zgc:112337; small nuclear ribon... 88.6 3e-17
hsa:11066 SNRNP35, HM-1, MGC138160, U1SNRNPBP; small nuclear r... 87.4 5e-17
xla:779322 snrnp35, MGC154877, u1snrnpbp; small nuclear ribonu... 84.3 5e-16
sce:YIL061C SNP1; Snp1p; K11093 U1 small nuclear ribonucleopro... 75.1 3e-13
mmu:101214 Tra2a, 1500010G04Rik, AL022798, G430041M01Rik, mAWM... 70.5 6e-12
hsa:29896 TRA2A, HSU53209; transformer 2 alpha homolog (Drosop... 70.5 6e-12
xla:379908 tra2a, MGC53025; transformer 2 alpha homolog; K1289... 68.6 3e-11
dre:393388 tra2a, MGC64175, zgc:64175; transformer-2 alpha; K1... 67.4 6e-11
xla:444408 MGC82977 protein; K12897 transformer-2 protein 65.5 2e-10
mmu:20462 Tra2b, 5730405G21Rik, D16Ertd266e, MGC102169, SIG-41... 63.9 7e-10
hsa:6434 TRA2B, DKFZp686F18120, Htra2-beta, SFRS10, SRFS10, TR... 63.9 7e-10
ath:AT3G46020 RNA-binding protein, putative 63.9 7e-10
hsa:1153 CIRBP, CIRP; cold inducible RNA binding protein; K131... 63.2 1e-09
mmu:12696 Cirbp, Cirp, R74941; cold inducible RNA binding prot... 62.8 1e-09
dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cyt... 62.0 2e-09
hsa:10929 SRSF8, SFRS2B, SRP46; serine/arginine-rich splicing ... 61.6 3e-09
hsa:378951 RBMY1J; RNA binding motif protein, Y-linked, family... 61.2 4e-09
hsa:159163 RBMY1F, MGC33094, YRRM2; RNA binding motif protein,... 61.2 4e-09
xla:398448 tra2b, MGC132359, sfrs10; transformer 2 beta homolo... 61.2 4e-09
dre:550495 wu:fb12a12, wu:fb16h02, wu:fb55a06, wu:fc67h01, wu:... 61.2 4e-09
ath:AT3G23830 GRP4; GRP4 (GLYCINE-RICH RNA-BINDING PROTEIN 4);... 60.8 5e-09
mmu:67418 Ppil4, 3732410E19Rik, 3830425H19Rik, AI788954, AW146... 60.5 6e-09
ath:AT3G20930 RNA recognition motif (RRM)-containing protein 60.5 7e-09
dre:394172 tra2b, MGC56612, sfrs10, zgc:56612; transformer 2 b... 60.1 9e-09
xla:379484 cirbp-b, MGC64461, Xcirp, cirbp, cirp, cirp2, xcirp... 60.1 1e-08
hsa:85313 PPIL4, HDCME13P; peptidylprolyl isomerase (cyclophil... 60.1 1e-08
hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A) ... 59.3 1e-08
ath:AT2G27330 RNA recognition motif (RRM)-containing protein 58.9 2e-08
xla:379761 cirbp-a, MGC52663, XCIRP, XCIRP-1, cirbp, cirp, cir... 58.9 2e-08
dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09... 58.5 2e-08
hsa:5940 RBMY1A1, MGC181956, RBM1, RBM2, RBMY, RBMY1C, YRRM1, ... 58.5 3e-08
hsa:378950 RBMY1E; RNA binding motif protein, Y-linked, family... 58.5 3e-08
hsa:378949 RBMY1D; RNA binding motif protein, Y-linked, family... 58.5 3e-08
xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) bindin... 58.5 3e-08
hsa:378948 RBMY1B; RNA binding motif protein, Y-linked, family... 58.5 3e-08
ath:AT4G39260 GR-RBP8; RNA binding / nucleic acid binding / nu... 58.2 3e-08
ath:AT3G26420 ATRZ-1A; RNA binding / nucleotide binding 57.8 4e-08
> tgo:TGME49_005180 U1 small nuclear ribonucleoprotein, putative
; K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=274
Score = 267 bits (683), Expect = 3e-71, Method: Compositional matrix adjust.
Identities = 127/187 (67%), Positives = 148/187 (79%), Gaps = 0/187 (0%)
Query 95 MAAIGMPPHILALFQAREPLAPLPALKAKQPRPYAGVAEYLKKFESGPPPAASAFESPKQ 154
MAAIGMPPHILALFQAR PL P PA K K R Y G+A+Y+ KFESGPPP +E+PKQ
Sbjct 1 MAAIGMPPHILALFQARPPLDPFPAFKKKHTRGYVGLADYVNKFESGPPPPTVPYETPKQ 60
Query 155 RQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRREFEV 214
R R KR +LL HL +Q+ + Y P ED ++ GDPF TLFVG +SY+T+EKKL+REFE
Sbjct 61 RLLRQKRARLLSHLSKQKELIKQYNPKEDKKLTGDPFRTLFVGGISYDTTEKKLKREFEQ 120
Query 215 YGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERARTVPG 274
YG ++RVR++ D G+PRGY F+EFE DRDMKEAYK ADG KIDGRRVLVDVERARTVPG
Sbjct 121 YGSIKRVRLIYDRNGKPRGYGFIEFENDRDMKEAYKNADGKKIDGRRVLVDVERARTVPG 180
Query 275 WLPRRLG 281
WLPRRLG
Sbjct 181 WLPRRLG 187
> tpv:TP02_0544 U1 small nuclear ribonucleoprotein; K11093 U1
small nuclear ribonucleoprotein 70kDa
Length=227
Score = 200 bits (509), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 98/196 (50%), Positives = 136/196 (69%), Gaps = 9/196 (4%)
Query 95 MAAIGMPPHILALFQAREPLAPLPALKAKQPRPYAGVAEYLKKFESGPPPAASAFESPKQ 154
M+A+GMPPH+L LFQAR PL + L +K R G+A +L F++ PP FE+P+Q
Sbjct 1 MSALGMPPHLLVLFQARPPLQHVDPLPSKPVRQIDGLAGFLDSFDAETPPKKEPFETPRQ 60
Query 155 RQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRI---------QGDPFATLFVGKLSYETSE 205
R+ + KR+ LL + ++ +++A Y P DPR+ DP+ TLFV ++Y+ +E
Sbjct 61 RRDKRKRETLLKYKEELKKRASEYDPSYDPRLARGGTSSWLTHDPYRTLFVANIAYDVTE 120
Query 206 KKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVD 265
K+L +EF+ YG +RRVR++ D +PRGYAF+E+E +RDM AYK+ADG KI GRRV+VD
Sbjct 121 KQLSKEFQTYGKIRRVRMIHDRSNKPRGYAFIEYENERDMVTAYKRADGKKISGRRVIVD 180
Query 266 VERARTVPGWLPRRLG 281
VERARTV GWLPRRLG
Sbjct 181 VERARTVEGWLPRRLG 196
> bbo:BBOV_II005790 18.m06481; u1 snRNP; K11093 U1 small nuclear
ribonucleoprotein 70kDa
Length=247
Score = 194 bits (493), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 93/196 (47%), Positives = 132/196 (67%), Gaps = 9/196 (4%)
Query 95 MAAIGMPPHILALFQAREPLAPLPALKAKQPRPYAGVAEYLKKFESGPPPAASAFESPKQ 154
M+A+GMPPH+L LFQAR PL +P ++ + + G+A+Y+ F P FE+P Q
Sbjct 1 MSALGMPPHLLVLFQARPPLEYIPPIENGMKKKFNGIADYVGYFSKEQVPPDPPFETPHQ 60
Query 155 RQQRLKRQKLLLHLQQQQRQAEAYRPLEDP---------RIQGDPFATLFVGKLSYETSE 205
R + KR++++ H ++Q+ + +AY P DP + DP+ TLFV + YE +E
Sbjct 61 RADKKKRERIIEHHKKQKAERDAYDPKYDPALARGATNPWLTHDPYKTLFVSNIPYEVTE 120
Query 206 KKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVD 265
K+L +EF+VYG VRR+R++ D + PRGYAF+E+ +RDM AYK+ADG K+ GRRVLVD
Sbjct 121 KQLWKEFDVYGRVRRIRMINDRKNIPRGYAFIEYNDERDMVNAYKRADGRKVSGRRVLVD 180
Query 266 VERARTVPGWLPRRLG 281
VERARTV GW P+RLG
Sbjct 181 VERARTVAGWYPKRLG 196
> ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTEIN-70K);
RNA binding / nucleic acid binding / nucleotide binding;
K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=427
Score = 175 bits (444), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 94/220 (42%), Positives = 133/220 (60%), Gaps = 5/220 (2%)
Query 67 LRRSNAPPRCRCCCCRCCCCCGVYVQRKMAAIGMPPHILALFQAREPLAPLPALKAKQPR 126
LR NA + R + + + G+ ++L LF+ R PL P + ++
Sbjct 9 LRNPNAAVQARAKVQNRANVLQLKLMGQSHPTGLTNNLLKLFEPRPPLEYKPPPEKRKCP 68
Query 127 PYAGVAEYLKKF-ESGPP---PAASAFESPKQRQQRLKRQKLLLHLQQQQRQAEAYRPLE 182
PY G+A+++ F E G P P E P Q+++R+ + +L +++ + Y P
Sbjct 69 PYTGMAQFVSNFAEPGDPEYAPPKPEVELPSQKRERIHKLRLEKGVEKAAEDLKKYDPNN 128
Query 183 DPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDP-RGRPRGYAFVEFER 241
DP GDP+ TLFV +L+YE+SE K++REFE YGP++RV +V D +P+GYAF+E+
Sbjct 129 DPNATGDPYKTLFVSRLNYESSESKIKREFESYGPIKRVHLVTDQLTNKPKGYAFIEYMH 188
Query 242 DRDMKEAYKKADGTKIDGRRVLVDVERARTVPGWLPRRLG 281
RDMK AYK+ADG KIDGRRVLVDVER RTVP W PRRLG
Sbjct 189 TRDMKAAYKQADGQKIDGRRVLVDVERGRTVPNWRPRRLG 228
> pfa:MAL13P1.338 U1 small nuclear ribonucleoprotein, putative;
K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=423
Score = 171 bits (432), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 87/187 (46%), Positives = 129/187 (68%), Gaps = 0/187 (0%)
Query 95 MAAIGMPPHILALFQAREPLAPLPALKAKQPRPYAGVAEYLKKFESGPPPAASAFESPKQ 154
M+AIGMP HIL LFQAR L +K K+P+ Y+G++++L FE G P ES K+
Sbjct 1 MSAIGMPQHILILFQARPMLEFYKPIKKKKPKEYSGLSDFLNYFEEGEAPPKIKVESFKE 60
Query 155 RQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRREFEV 214
R+++ K++K+ + + + + Y P ++ + DP TLF+G+LSYE SE+KL++EFE
Sbjct 61 RKEKKKKEKMAYNELILKEKRKEYDPFKNEDLTSDPKKTLFIGRLSYEVSEQKLKKEFES 120
Query 215 YGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERARTVPG 274
YG ++ V+++ D +PRGYAF+EFE + M +AYK ADG KI+ RR+LVD+ER RT+
Sbjct 121 YGKIKTVKIIYDKNLKPRGYAFIEFEHTKSMNDAYKLADGKKIENRRILVDIERGRTIKN 180
Query 275 WLPRRLG 281
W+PRRLG
Sbjct 181 WIPRRLG 187
> mmu:20637 Snrnp70, 2700022N21Rik, 3200002N22Rik, 70kDa, AI325098,
R74807, Rnulp70, Snrp70, Srnp70, U1-70, U1-70K; small
nuclear ribonucleoprotein 70 (U1); K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=448
Score = 164 bits (416), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 90/185 (48%), Positives = 125/185 (67%), Gaps = 11/185 (5%)
Query 100 MPPHILALFQAREPLAPLPALKA-----KQPRPYAGVAEYLKKFES---GPPPAASAFES 151
+PP++LALF R+P+ LP L+ +PY G+A Y+++FE PPP +
Sbjct 5 LPPNLLALFAPRDPIPYLPPLEKLPHEKHHNQPYCGIAPYIREFEDPRDAPPPTRAETRE 64
Query 152 PKQRQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRRE 211
+R +R +R+K+ Q+ + + + + P DP QGD F TLFV +++Y+T+E KLRRE
Sbjct 65 --ERMERKRREKIERRQQEVETELKMWDPHNDPNAQGDAFKTLFVARVNYDTTESKLRRE 122
Query 212 FEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERAR 270
FEVYGP++R+ +V R G+PRGYAF+E+E +RDM AYK ADG KIDGRRVLVDVER R
Sbjct 123 FEVYGPIKRIHMVYSKRSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRRVLVDVERGR 182
Query 271 TVPGW 275
TV GW
Sbjct 183 TVKGW 187
> hsa:6625 SNRNP70, RNPU1Z, RPU1, SNRP70, Snp1, U1-70K, U170K,
U1AP, U1RNP; small nuclear ribonucleoprotein 70kDa (U1); K11093
U1 small nuclear ribonucleoprotein 70kDa
Length=437
Score = 164 bits (416), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 90/185 (48%), Positives = 125/185 (67%), Gaps = 11/185 (5%)
Query 100 MPPHILALFQAREPLAPLPALKA-----KQPRPYAGVAEYLKKFES---GPPPAASAFES 151
+PP++LALF R+P+ LP L+ +PY G+A Y+++FE PPP +
Sbjct 5 LPPNLLALFAPRDPIPYLPPLEKLPHEKHHNQPYCGIAPYIREFEDPRDAPPPTRAETRE 64
Query 152 PKQRQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRRE 211
+R +R +R+K+ Q+ + + + + P DP QGD F TLFV +++Y+T+E KLRRE
Sbjct 65 --ERMERKRREKIERRQQEVETELKMWDPHNDPNAQGDAFKTLFVARVNYDTTESKLRRE 122
Query 212 FEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERAR 270
FEVYGP++R+ +V R G+PRGYAF+E+E +RDM AYK ADG KIDGRRVLVDVER R
Sbjct 123 FEVYGPIKRIHMVYSKRSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRRVLVDVERGR 182
Query 271 TVPGW 275
TV GW
Sbjct 183 TVKGW 187
> cel:K04G7.10 rnp-7; RNP (RRM RNA binding domain) containing
family member (rnp-7); K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=332
Score = 163 bits (412), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 85/189 (44%), Positives = 121/189 (64%), Gaps = 7/189 (3%)
Query 100 MPPHILALFQAREPLAPLP------ALKAKQPRPYAGVAEYLKKFES-GPPPAASAFESP 152
+PP++LALF+AR P+ LP K + P GVA+Y++ FE PA +
Sbjct 5 LPPNLLALFEARPPVQYLPPCQDLLVDKNAKRAPMTGVAQYIQLFEDPKDTPAKIPVVTK 64
Query 153 KQRQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRREF 212
+ ++ +RQK L + ++ + P E+PR DP+ TLFVG+++YETSE KLRREF
Sbjct 65 AEAKEEKRRQKDELLAYKVEQGIATWNPAENPRASEDPYRTLFVGRINYETSESKLRREF 124
Query 213 EVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERARTV 272
E YG ++++ +V D G+PRGYAF+E+ +M AYKKADG K+DG+R++VD ER RT
Sbjct 125 EAYGKIKKLTMVHDEAGKPRGYAFIEYSDKAEMHTAYKKADGIKVDGKRLVVDYERGRTQ 184
Query 273 PGWLPRRLG 281
WLPRRLG
Sbjct 185 KTWLPRRLG 193
> xla:397946 snrnp70, MGC130688, snrp70; small nuclear ribonucleoprotein
70kDa (U1); K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=471
Score = 161 bits (408), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 85/188 (45%), Positives = 121/188 (64%), Gaps = 14/188 (7%)
Query 100 MPPHILALFQAREPLAPLPAL-----KAKQPRPYAGVAEYLKKFES---GPPPAASAFES 151
+PP++LALF R+P+ LP L + +PY G+A Y+++FE PPP +
Sbjct 5 LPPNLLALFAPRDPVPYLPPLDKLPHEKHHNQPYCGIAPYIREFEDPRDAPPPTRAETRE 64
Query 152 PKQRQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRRE 211
+ ++R ++ + Q + + + + P D QGD F TLFV +++Y+T+E KLRRE
Sbjct 65 ERMERKRREKIERRQ--QDVENELKIWDPHNDQNAQGDAFKTLFVARVNYDTTESKLRRE 122
Query 212 FEVYGPVRRVRVVRDP----RGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVE 267
FEVYGP++R+ +V + G+PRGYAF+E+E +RDM AYK ADG KIDGRRVLVDVE
Sbjct 123 FEVYGPIKRIHIVYNKGSEGSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRRVLVDVE 182
Query 268 RARTVPGW 275
R RTV GW
Sbjct 183 RGRTVKGW 190
> dre:445398 snrnp70, MGC136450, snrp70, wu:fc20b04, zgc:136450;
small nuclear ribonucleoprotein 70 (U1); K11093 U1 small
nuclear ribonucleoprotein 70kDa
Length=495
Score = 157 bits (397), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 83/185 (44%), Positives = 121/185 (65%), Gaps = 11/185 (5%)
Query 100 MPPHILALFQAREPLAPLPAL-----KAKQPRPYAGVAEYLKKFES---GPPPAASAFES 151
+PP++LALF R+P+ LP L + +PY G+A +++ FE PPP +
Sbjct 5 LPPNLLALFAPRDPIPFLPQLGKLPHEKHHNQPYCGIAPFIRHFEDPRDAPPPTRAETRE 64
Query 152 PKQRQQRLKRQKLLLHLQQQQRQAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRRE 211
+ ++R ++ + + + + + + P D QGD F TLFV +++Y+T+E KLRRE
Sbjct 65 ERLERKRREKMERRQVVVEGELKL--WDPHNDANAQGDAFKTLFVARINYDTTESKLRRE 122
Query 212 FEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERAR 270
FEVYGP++R+ +V + + G+PRGYAF+E+E +RDM AYK ADG KIDGRRVLVDVER R
Sbjct 123 FEVYGPIKRIYIVYNKKTGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRRVLVDVERGR 182
Query 271 TVPGW 275
TV GW
Sbjct 183 TVKGW 187
> cpv:cgd6_990 U1 snrp Snp1p. RRM domain containing protein
Length=281
Score = 157 bits (396), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 80/196 (40%), Positives = 123/196 (62%), Gaps = 13/196 (6%)
Query 95 MAAIGMPPHILALFQAREPLAPLPALKAKQPRPYAGVAEYLK-------KFESGPPPAAS 147
M+A+GM +L FQAR PL +P + +PY G+A+ + K + P P
Sbjct 1 MSAVGMSNDLLKFFQARPPLPYVPYPNRRPHKPYQGLADLINENPDLFDKVNTPPEP--- 57
Query 148 AFESPKQRQQRLKRQKLLLHLQQQQRQAEAYRPLED--PRIQGDPFATLFVGKLSYETSE 205
+ S K R++ K Q++ +++ +++ E+Y P+E+ DP+ TLF+ +L+Y+T+E
Sbjct 58 -YISKKMRRETEKLQRIQNYMKDMRKKIESYNPMEENVDNKTFDPYNTLFIARLNYDTTE 116
Query 206 KKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLVD 265
+ L+RE EVYG + +R+V+D +G RGYAFVEF+ + +KEAYK + IDG +VLVD
Sbjct 117 RTLKRELEVYGKILNLRIVKDFQGESRGYAFVEFDNEDSLKEAYKSFNKKSIDGWKVLVD 176
Query 266 VERARTVPGWLPRRLG 281
VER RTV WLP+RLG
Sbjct 177 VERGRTVENWLPKRLG 192
> ath:AT2G43370 U1 small nuclear ribonucleoprotein 70 kDa, putative;
K13155 U11/U12 small nuclear ribonucleoprotein 35 kDa
protein
Length=333
Score = 96.7 bits (239), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/105 (43%), Positives = 66/105 (62%), Gaps = 1/105 (0%)
Query 174 QAEAYRPLEDPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPR 232
A Y P D + GDP+ TLFVG+LS+ T+E LR YG ++ +R+VR G R
Sbjct 46 NAGLYDPSGDSKAVGDPYCTLFVGRLSHHTTEDTLREVMSKYGRIKNLRLVRHIVTGASR 105
Query 233 GYAFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERARTVPGWLP 277
GY FVE+E +++M AY+ A + IDGR ++VD R + +PGW+P
Sbjct 106 GYGFVEYETEKEMLRAYEDAHHSLIDGREIIVDYNRQQLMPGWIP 150
> mmu:76167 Snrnp35, 6330548G22Rik; small nuclear ribonucleoprotein
35 (U11/U12); K13155 U11/U12 small nuclear ribonucleoprotein
35 kDa protein
Length=244
Score = 89.7 bits (221), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 43/93 (46%), Positives = 63/93 (67%), Gaps = 1/93 (1%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRD 244
+ GDP TLFV +L+ +T E+KL+ F YG +RR+R+VRD G +GYAF+E++ +R
Sbjct 45 VTGDPLLTLFVARLNLQTKEEKLKEVFSRYGDIRRLRLVRDLVTGFSKGYAFIEYKEERA 104
Query 245 MKEAYKKADGTKIDGRRVLVDVERARTVPGWLP 277
+ +AY+ ADG ID + VD E RT+ GW+P
Sbjct 105 LMKAYRDADGLVIDQHEIFVDYELERTLRGWIP 137
> dre:569999 snrnp35, MGC112337, zgc:112337; small nuclear ribonucleoprotein
35 (U11/U12); K13155 U11/U12 small nuclear ribonucleoprotein
35 kDa protein
Length=208
Score = 88.6 bits (218), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 45/94 (47%), Positives = 67/94 (71%), Gaps = 1/94 (1%)
Query 188 GDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDP-RGRPRGYAFVEFERDRDMK 246
GDP TLFV +L+ +T+E+KLR F +G +RR+R+VRD G + YAF+E++ +R +K
Sbjct 46 GDPDLTLFVARLNPQTTEEKLRDVFSKFGDIRRLRLVRDVVTGFSKRYAFIEYKEERSLK 105
Query 247 EAYKKADGTKIDGRRVLVDVERARTVPGWLPRRL 280
A++ A+ +D +LVDVE+ RT+PGW PRRL
Sbjct 106 RAWRDANKLILDQYELLVDVEQERTLPGWRPRRL 139
> hsa:11066 SNRNP35, HM-1, MGC138160, U1SNRNPBP; small nuclear
ribonucleoprotein 35kDa (U11/U12); K13155 U11/U12 small nuclear
ribonucleoprotein 35 kDa protein
Length=246
Score = 87.4 bits (215), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 43/93 (46%), Positives = 62/93 (66%), Gaps = 1/93 (1%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRD 244
+ GDP TLFV +L+ +T E KL+ F YG +RR+R+VRD G +GYAF+E++ +R
Sbjct 45 VIGDPLLTLFVARLNLQTKEDKLKEVFSRYGDIRRLRLVRDLVTGFSKGYAFIEYKEERA 104
Query 245 MKEAYKKADGTKIDGRRVLVDVERARTVPGWLP 277
+ +AY+ ADG ID + VD E RT+ GW+P
Sbjct 105 VIKAYRDADGLVIDQHEIFVDYELERTLKGWIP 137
> xla:779322 snrnp35, MGC154877, u1snrnpbp; small nuclear ribonucleoprotein
35kDa (U11/U12); K13155 U11/U12 small nuclear
ribonucleoprotein 35 kDa protein
Length=272
Score = 84.3 bits (207), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 40/93 (43%), Positives = 63/93 (67%), Gaps = 1/93 (1%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRD 244
+ GDP TLFV +LS +T+E+KL+ F YG ++R+R+VRD G +GYAF+E++++
Sbjct 45 VTGDPHLTLFVSRLSPQTTEEKLKEVFSRYGDIKRIRLVRDFITGFSKGYAFIEYKQENA 104
Query 245 MKEAYKKADGTKIDGRRVLVDVERARTVPGWLP 277
+ +A++ A+ ID R V VD E R + GW+P
Sbjct 105 IMKAHRDANKLVIDQREVFVDFELERNLKGWIP 137
> sce:YIL061C SNP1; Snp1p; K11093 U1 small nuclear ribonucleoprotein
70kDa
Length=300
Score = 75.1 bits (183), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 59/197 (29%), Positives = 97/197 (49%), Gaps = 29/197 (14%)
Query 101 PPHILALFQAREPLA-----PLPALKAKQPRPYAGVAE--------YLKKFESGPPPAAS 147
P + LF+ R PL+ P K + GVA Y+++F G
Sbjct 9 PDDVSRLFKPRPPLSYKRPTDYPYAKRQTNPNITGVANLLSTSLKHYMEEFPEG------ 62
Query 148 AFESPKQRQQRLKRQKL--LLHLQQQQRQAEAYRPLEDPRIQG-DPFATLFVGKLSYETS 204
SP QR + KL + + Q R+ + + P DP I+ DP+ T+F+G+L Y+
Sbjct 63 ---SPNNHLQRYEDIKLSKIKNAQLLDRRLQNWNPNVDPHIKDTDPYRTIFIGRLPYDLD 119
Query 205 EKKLRREFEVYGPVRRVRVVRDP-RGRPRGYAFVEFERDRDMKEAYKKAD---GTKIDGR 260
E +L++ F +G + ++R+V+D + +GYAF+ F+ K A+K+ G +I R
Sbjct 120 EIELQKYFVKFGEIEKIRIVKDKITQKSKGYAFIVFKDPISSKMAFKEIGVHRGIQIKDR 179
Query 261 RVLVDVERARTVPGWLP 277
+VD+ER RTV + P
Sbjct 180 ICIVDIERGRTVKYFKP 196
> mmu:101214 Tra2a, 1500010G04Rik, AL022798, G430041M01Rik, mAWMS1;
transformer 2 alpha homolog (Drosophila); K12897 transformer-2
protein
Length=282
Score = 70.5 bits (171), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 47/95 (49%), Positives = 58/95 (61%), Gaps = 5/95 (5%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDR 243
R DP L V LS T+E+ LR F YGP+ V VV D R GR RG+AFV FER
Sbjct 110 RANPDPNTCLGVFGLSLYTTERDLREVFSRYGPLSGVNVVYDQRTGRSRGFAFVYFERID 169
Query 244 DMKEAYKKADGTKIDGRRVLVD---VERART-VPG 274
D KEA ++A+G ++DGRR+ VD +RA T PG
Sbjct 170 DSKEAMERANGMELDGRRIRVDYSITKRAHTPTPG 204
> hsa:29896 TRA2A, HSU53209; transformer 2 alpha homolog (Drosophila);
K12897 transformer-2 protein
Length=282
Score = 70.5 bits (171), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 47/95 (49%), Positives = 58/95 (61%), Gaps = 5/95 (5%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDR 243
R DP L V LS T+E+ LR F YGP+ V VV D R GR RG+AFV FER
Sbjct 112 RANPDPNTCLGVFGLSLYTTERDLREVFSRYGPLSGVNVVYDQRTGRSRGFAFVYFERID 171
Query 244 DMKEAYKKADGTKIDGRRVLVD---VERART-VPG 274
D KEA ++A+G ++DGRR+ VD +RA T PG
Sbjct 172 DSKEAMERANGMELDGRRIRVDYSITKRAHTPTPG 206
> xla:379908 tra2a, MGC53025; transformer 2 alpha homolog; K12897
transformer-2 protein
Length=276
Score = 68.6 bits (166), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 45/95 (47%), Positives = 57/95 (60%), Gaps = 5/95 (5%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDR 243
R DP + V LS T+E+ +R F YGP+ V VV D R GR RG+AFV FER
Sbjct 105 RANPDPNLCIGVFGLSLYTTERDIREVFSRYGPLAGVNVVYDQRTGRSRGFAFVYFERIE 164
Query 244 DMKEAYKKADGTKIDGRRVLVD---VERART-VPG 274
D +EA + ADG ++DGRR+ VD +RA T PG
Sbjct 165 DSREAMEHADGMELDGRRIRVDYSITKRAHTPTPG 199
> dre:393388 tra2a, MGC64175, zgc:64175; transformer-2 alpha;
K12897 transformer-2 protein
Length=297
Score = 67.4 bits (163), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 41/85 (48%), Positives = 51/85 (60%), Gaps = 1/85 (1%)
Query 183 DPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPR-GRPRGYAFVEFER 241
D R DP L V LS T+E+ LR F YG + V VV D R GR RG+AFV FE
Sbjct 126 DARANPDPNTCLGVFGLSLYTTERDLREVFSRYGSLAGVNVVYDQRTGRSRGFAFVYFEH 185
Query 242 DRDMKEAYKKADGTKIDGRRVLVDV 266
D KEA ++A+G ++DGRR+ VD
Sbjct 186 IDDAKEAMERANGMELDGRRIRVDY 210
> xla:444408 MGC82977 protein; K12897 transformer-2 protein
Length=276
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 43/95 (45%), Positives = 56/95 (58%), Gaps = 5/95 (5%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDR 243
R DP + V LS T+E+ LR F YGP+ V VV D R GR RG+AFV FER
Sbjct 105 RANPDPNICVGVFGLSLYTTERDLREVFSRYGPLSSVNVVYDQRTGRSRGFAFVYFERME 164
Query 244 DMKEAYKKADGTKIDGRRVLVD---VERART-VPG 274
D +EA + +G ++DGR++ VD +RA T PG
Sbjct 165 DSREAMEHVNGMELDGRKLRVDYSITKRAHTPTPG 199
> mmu:20462 Tra2b, 5730405G21Rik, D16Ertd266e, MGC102169, SIG-41,
Sfrs10, Silg41, TRA2beta; transformer 2 beta homolog (Drosophila);
K12897 transformer-2 protein
Length=288
Score = 63.9 bits (154), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 38/83 (45%), Positives = 50/83 (60%), Gaps = 1/83 (1%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGR-PRGYAFVEFERDR 243
R DP L V LS T+E+ LR F YGP+ V +V D + R RG+AFV FE
Sbjct 111 RANPDPNCCLGVFGLSLYTTERDLREVFSKYGPIADVSIVYDQQSRRSRGFAFVYFENVD 170
Query 244 DMKEAYKKADGTKIDGRRVLVDV 266
D KEA ++A+G ++DGRR+ VD
Sbjct 171 DAKEAKERANGMELDGRRIRVDF 193
> hsa:6434 TRA2B, DKFZp686F18120, Htra2-beta, SFRS10, SRFS10,
TRA2-BETA, TRAN2B; transformer 2 beta homolog (Drosophila);
K12897 transformer-2 protein
Length=288
Score = 63.9 bits (154), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 38/83 (45%), Positives = 50/83 (60%), Gaps = 1/83 (1%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGR-PRGYAFVEFERDR 243
R DP L V LS T+E+ LR F YGP+ V +V D + R RG+AFV FE
Sbjct 111 RANPDPNCCLGVFGLSLYTTERDLREVFSKYGPIADVSIVYDQQSRRSRGFAFVYFENVD 170
Query 244 DMKEAYKKADGTKIDGRRVLVDV 266
D KEA ++A+G ++DGRR+ VD
Sbjct 171 DAKEAKERANGMELDGRRIRVDF 193
> ath:AT3G46020 RNA-binding protein, putative
Length=102
Score = 63.9 bits (154), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 49/76 (64%), Gaps = 1/76 (1%)
Query 192 ATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRG-RPRGYAFVEFERDRDMKEAYK 250
A LFV +LS T+++ LR+ F +G ++ R++RD RP+G+ F+ F+ + D ++A K
Sbjct 7 AQLFVSRLSAYTTDQSLRQLFSPFGQIKEARLIRDSETQRPKGFGFITFDSEDDARKALK 66
Query 251 KADGTKIDGRRVLVDV 266
DG +DGR + V+V
Sbjct 67 SLDGKIVDGRLIFVEV 82
> hsa:1153 CIRBP, CIRP; cold inducible RNA binding protein; K13195
cold-inducible RNA-binding protein
Length=172
Score = 63.2 bits (152), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 45/73 (61%), Gaps = 1/73 (1%)
Query 194 LFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRDMKEAYKKA 252
LFVG LS++T+E+ L + F YG + V VV+D R RG+ FV FE D K+A
Sbjct 8 LFVGGLSFDTNEQSLEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAM 67
Query 253 DGTKIDGRRVLVD 265
+G +DGR++ VD
Sbjct 68 NGKSVDGRQIRVD 80
> mmu:12696 Cirbp, Cirp, R74941; cold inducible RNA binding protein;
K13195 cold-inducible RNA-binding protein
Length=172
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 45/73 (61%), Gaps = 1/73 (1%)
Query 194 LFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRDMKEAYKKA 252
LFVG LS++T+E+ L + F YG + V VV+D R RG+ FV FE D K+A
Sbjct 8 LFVGGLSFDTNEQALEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAM 67
Query 253 DGTKIDGRRVLVD 265
+G +DGR++ VD
Sbjct 68 NGKSVDGRQIRVD 80
> dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cytoplasmic
4 (inducible form); K13126 polyadenylate-binding
protein
Length=637
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 191 FATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYK 250
F +++ + +++L+ F+ YG V+V+ DP G+ RG+ FV +E+ D +A +
Sbjct 191 FTNVYIKNFGDDMDDQRLKELFDKYGKTLSVKVMTDPTGKSRGFGFVSYEKHEDANKAVE 250
Query 251 KADGTKIDGRRVLV 264
+ +GT+++G+ V V
Sbjct 251 EMNGTELNGKTVFV 264
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 1/82 (1%)
Query 183 DPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERD 242
DP ++ +F+ L K L F +G + +VV D G +GYAFV FE
Sbjct 91 DPSLRKSGVGNVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGS-KGYAFVHFETQ 149
Query 243 RDMKEAYKKADGTKIDGRRVLV 264
A +K +G ++ R+V V
Sbjct 150 DAADRAIEKMNGMLLNDRKVFV 171
Score = 44.3 bits (103), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Query 190 PFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRG-RPRGYAFVEFERDRDMKEA 248
P A+L+VG L + +E L +F GPV +RV RD R GYA+V F++ D + A
Sbjct 10 PMASLYVGDLHPDITEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERA 69
Query 249 YKKADGTKIDGRRVLV 264
+ + G+ + +
Sbjct 70 LDTMNFDVVKGKPIRI 85
Score = 39.7 bits (91), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 26/92 (28%), Positives = 45/92 (48%), Gaps = 10/92 (10%)
Query 169 QQQQRQAEAYRPLED------PRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVR 222
++ +RQAE R E R QG L++ L ++KLR+EF +G + +
Sbjct 269 KKMERQAELKRKFEQLKQERISRYQG---VNLYIKNLDDTIDDEKLRKEFSPFGSITSAK 325
Query 223 VVRDPRGRPRGYAFVEFERDRDMKEAYKKADG 254
V+ + GR +G+ FV F + +A + +G
Sbjct 326 VMLE-EGRSKGFGFVCFSSPEEATKAVTEMNG 356
> hsa:10929 SRSF8, SFRS2B, SRP46; serine/arginine-rich splicing
factor 8; K12891 splicing factor, arginine/serine-rich 2
Length=282
Score = 61.6 bits (148), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 47/81 (58%), Gaps = 1/81 (1%)
Query 189 DPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGR-PRGYAFVEFERDRDMKE 247
D TL V L+Y TS LRR FE YG V V + R+P + PRG+AFV F RD ++
Sbjct 11 DGMITLKVDNLTYRTSPDSLRRVFEKYGRVGDVYIPREPHTKAPRGFAFVRFHDRRDAQD 70
Query 248 AYKKADGTKIDGRRVLVDVER 268
A DG ++DGR + V V R
Sbjct 71 AEAAMDGAELDGRELRVQVAR 91
> hsa:378951 RBMY1J; RNA binding motif protein, Y-linked, family
1, member J
Length=496
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 48/80 (60%), Gaps = 0/80 (0%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDM 245
++ D LF+G L+ ET+EK L+ F +GP+ V +++D + RG+AF+ FE D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 246 KEAYKKADGTKIDGRRVLVD 265
K A K +GT + G+ + V+
Sbjct 62 KNAAKDMNGTSLHGKAIKVE 81
> hsa:159163 RBMY1F, MGC33094, YRRM2; RNA binding motif protein,
Y-linked, family 1, member F
Length=496
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 48/80 (60%), Gaps = 0/80 (0%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDM 245
++ D LF+G L+ ET+EK L+ F +GP+ V +++D + RG+AF+ FE D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 246 KEAYKKADGTKIDGRRVLVD 265
K A K +GT + G+ + V+
Sbjct 62 KNAAKDMNGTSLHGKAIKVE 81
> xla:398448 tra2b, MGC132359, sfrs10; transformer 2 beta homolog;
K12897 transformer-2 protein
Length=298
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 37/83 (44%), Positives = 50/83 (60%), Gaps = 1/83 (1%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGR-PRGYAFVEFERDR 243
R DP L V LS T+E+ LR F YGP+ V +V D + R RG++FV FE
Sbjct 115 RANPDPNCCLGVFGLSLYTTERDLREVFSKYGPLSDVSIVYDQQSRRSRGFSFVYFENVD 174
Query 244 DMKEAYKKADGTKIDGRRVLVDV 266
D KEA ++A+G ++DGRR+ VD
Sbjct 175 DAKEAKERANGMELDGRRLRVDF 197
> dre:550495 wu:fb12a12, wu:fb16h02, wu:fb55a06, wu:fc67h01, wu:fj10g10,
zgc:109887; zgc:112425; K13195 cold-inducible RNA-binding
protein
Length=185
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query 194 LFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRDMKEAYKKA 252
LF+G LS++T+E+ L F YG + V V R+ R RG+ FV FE D K+A +
Sbjct 7 LFIGGLSFDTTEQSLEDAFSKYGVITNVHVARNRETNRSRGFGFVTFENPDDAKDALEGM 66
Query 253 DGTKIDGRRVLVD 265
+G +DGR + VD
Sbjct 67 NGKSVDGRTIRVD 79
> ath:AT3G23830 GRP4; GRP4 (GLYCINE-RICH RNA-BINDING PROTEIN 4);
RNA binding / double-stranded DNA binding / single-stranded
DNA binding
Length=136
Score = 60.8 bits (146), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 45/79 (56%), Gaps = 1/79 (1%)
Query 194 LFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRDMKEAYKKA 252
LFVG LS+ T + L++ F +G V V+ D GR RG+ FV F + A K+
Sbjct 37 LFVGGLSWGTDDSSLKQAFTSFGEVTEATVIADRETGRSRGFGFVSFSCEDSANNAIKEM 96
Query 253 DGTKIDGRRVLVDVERART 271
DG +++GR++ V++ R+
Sbjct 97 DGKELNGRQIRVNLATERS 115
> mmu:67418 Ppil4, 3732410E19Rik, 3830425H19Rik, AI788954, AW146233,
PPIase; peptidylprolyl isomerase (cyclophilin)-like 4
(EC:5.2.1.8); K12735 peptidyl-prolyl cis-trans isomerase-like
4 [EC:5.2.1.8]
Length=492
Score = 60.5 bits (145), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/81 (39%), Positives = 49/81 (60%), Gaps = 1/81 (1%)
Query 190 PFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDRDMKEA 248
P LFV KL+ T+++ L F +GP+R V+RD + G YAF+EFE++ D ++A
Sbjct 238 PENVLFVCKLNPVTTDEDLEIIFSRFGPIRSCEVIRDWKTGESLCYAFIEFEKEEDCEKA 297
Query 249 YKKADGTKIDGRRVLVDVERA 269
+ K D ID RR+ VD ++
Sbjct 298 FFKMDNVLIDDRRIHVDFSQS 318
> ath:AT3G20930 RNA recognition motif (RRM)-containing protein
Length=374
Score = 60.5 bits (145), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 35/96 (36%), Positives = 54/96 (56%), Gaps = 7/96 (7%)
Query 182 EDPRIQGD----PFAT--LFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRG-RPRGY 234
+D R Q D P T LF+ LS+ TSEK LR FE +G + V+++ D R +GY
Sbjct 266 QDSRDQDDSESPPVKTKKLFITGLSFYTSEKTLRAAFEGFGELVEVKIIMDKISKRSKGY 325
Query 235 AFVEFERDRDMKEAYKKADGTKIDGRRVLVDVERAR 270
AF+E+ + A K+ +G I+G ++VDV + +
Sbjct 326 AFLEYTTEEAAGTALKEMNGKIINGWMIVVDVAKTK 361
> dre:394172 tra2b, MGC56612, sfrs10, zgc:56612; transformer 2
beta homolog (Drosophila); K12897 transformer-2 protein
Length=278
Score = 60.1 bits (144), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 37/83 (44%), Positives = 49/83 (59%), Gaps = 1/83 (1%)
Query 185 RIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGR-PRGYAFVEFERDR 243
R DP L V LS T+E+ LR F YGP+ V +V D + R RG+A V FE
Sbjct 116 RANPDPNCCLGVFGLSLYTTERDLREVFSKYGPLSDVCIVYDQQSRRSRGFALVYFENRE 175
Query 244 DMKEAYKKADGTKIDGRRVLVDV 266
D KEA ++A+G ++DGRR+ VD
Sbjct 176 DSKEAKERANGMELDGRRIRVDY 198
> xla:379484 cirbp-b, MGC64461, Xcirp, cirbp, cirp, cirp2, xcirp2;
cold inducible RNA binding protein; K13195 cold-inducible
RNA-binding protein
Length=166
Score = 60.1 bits (144), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 45/73 (61%), Gaps = 1/73 (1%)
Query 194 LFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRDMKEAYKKA 252
LF+G L+++T+E+ L + F YG + V VV+D R RG+ FV FE D K+A
Sbjct 7 LFIGGLNFDTNEESLEQVFSKYGQISEVVVVKDRETKRSRGFGFVTFENPDDAKDAMMAM 66
Query 253 DGTKIDGRRVLVD 265
+G +DGR++ VD
Sbjct 67 NGKAVDGRQIRVD 79
> hsa:85313 PPIL4, HDCME13P; peptidylprolyl isomerase (cyclophilin)-like
4 (EC:5.2.1.8); K12735 peptidyl-prolyl cis-trans
isomerase-like 4 [EC:5.2.1.8]
Length=492
Score = 60.1 bits (144), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/81 (39%), Positives = 49/81 (60%), Gaps = 1/81 (1%)
Query 190 PFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPR-GRPRGYAFVEFERDRDMKEA 248
P LFV KL+ T+++ L F +GP+R V+RD + G YAF+EFE++ D ++A
Sbjct 238 PENVLFVCKLNPVTTDEDLEIIFSRFGPIRSCEVIRDWKTGESLCYAFIEFEKEEDCEKA 297
Query 249 YKKADGTKIDGRRVLVDVERA 269
+ K D ID RR+ VD ++
Sbjct 298 FFKMDNVLIDDRRIHVDFSQS 318
> hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A)
binding protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=660
Score = 59.3 bits (142), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 42/74 (56%), Gaps = 0/74 (0%)
Query 191 FATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYK 250
F +++ E ++ L+ F +G V+V+RDP G+ +G+ FV +E+ D +A +
Sbjct 190 FTNVYIKNFGEEVDDESLKELFSQFGKTLSVKVMRDPNGKSKGFGFVSYEKHEDANKAVE 249
Query 251 KADGTKIDGRRVLV 264
+ +G +I G+ + V
Sbjct 250 EMNGKEISGKIIFV 263
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 1/82 (1%)
Query 183 DPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERD 242
DP ++ +F+ L K L F +G + +VV D G +GYAFV FE
Sbjct 90 DPSLRKSGVGNVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGS-KGYAFVHFETQ 148
Query 243 RDMKEAYKKADGTKIDGRRVLV 264
+A +K +G ++ R+V V
Sbjct 149 EAADKAIEKMNGMLLNDRKVFV 170
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Query 190 PFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRG-RPRGYAFVEFERDRDMKEA 248
P A+L+VG L + +E L +F GPV +RV RD R GYA+V F++ D + A
Sbjct 9 PMASLYVGDLHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERA 68
Query 249 YKKADGTKIDGRRVLV 264
+ I G+ + +
Sbjct 69 LDTMNFDVIKGKPIRI 84
Score = 39.7 bits (91), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 47/99 (47%), Gaps = 10/99 (10%)
Query 172 QRQAEAYRPLED------PRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVR 225
+RQAE R E R QG L++ L ++KLR+EF +G + +V+
Sbjct 271 ERQAELKRKFEQLKQERISRYQG---VNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVML 327
Query 226 DPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLV 264
+ GR +G+ FV F + +A + +G + + + V
Sbjct 328 E-DGRSKGFGFVCFSSPEEATKAVTEMNGRIVGSKPLYV 365
> ath:AT2G27330 RNA recognition motif (RRM)-containing protein
Length=116
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 49/79 (62%), Gaps = 1/79 (1%)
Query 192 ATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDP-RGRPRGYAFVEFERDRDMKEAYK 250
+TLFV +S+ ++E+ L + F YG V +V V+ D R RP+G+A+V F + ++A
Sbjct 21 STLFVKGISFSSTEETLTQAFSQYGQVLKVDVIMDKIRCRPKGFAYVTFSSKEEAEKALL 80
Query 251 KADGTKIDGRRVLVDVERA 269
+ + +DGR V++D +A
Sbjct 81 ELNAQLVDGRVVILDTTKA 99
> xla:379761 cirbp-a, MGC52663, XCIRP, XCIRP-1, cirbp, cirp, cirp-1;
cold inducible RNA binding protein
Length=166
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 44/73 (60%), Gaps = 1/73 (1%)
Query 194 LFVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRDMKEAYKKA 252
LF+G L++ET+E L + F YG + V VV+D R RG+ FV FE D K+A
Sbjct 7 LFIGGLNFETNEDCLEQAFTKYGRISEVVVVKDRETKRSRGFGFVTFENVDDAKDAMMAM 66
Query 253 DGTKIDGRRVLVD 265
+G +DGR++ VD
Sbjct 67 NGKSVDGRQIRVD 79
> dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09,
wu:fj61f06, zgc:109879; poly A binding protein, cytoplasmic
1 a; K13126 polyadenylate-binding protein
Length=634
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 43/74 (58%), Gaps = 0/74 (0%)
Query 191 FATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYK 250
F +++ + ++KL+ F YGP +RV+ D G+ +G+ FV FER D + A
Sbjct 190 FTNVYIKNFGEDMDDEKLKEIFCKYGPALSIRVMTDDSGKSKGFGFVSFERHEDAQRAVD 249
Query 251 KADGTKIDGRRVLV 264
+ +G +++G++V V
Sbjct 250 EMNGKEMNGKQVYV 263
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 42/83 (50%), Gaps = 1/83 (1%)
Query 183 DPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRG-RPRGYAFVEFER 241
+P P A+L+VG L + +E L +F GP+ +RV RD R GYA+V F++
Sbjct 2 NPSAPSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMMTRRSLGYAYVNFQQ 61
Query 242 DRDMKEAYKKADGTKIDGRRVLV 264
D + A + I GR V +
Sbjct 62 PADAERALDTMNFDVIKGRPVRI 84
Score = 47.4 bits (111), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 38/82 (46%), Gaps = 1/82 (1%)
Query 183 DPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERD 242
DP ++ +F+ L K L F +G + +VV D G +GY FV FE
Sbjct 90 DPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGS-KGYGFVHFETH 148
Query 243 RDMKEAYKKADGTKIDGRRVLV 264
+ A +K +G ++ R+V V
Sbjct 149 EAAERAIEKMNGMLLNDRKVFV 170
Score = 35.4 bits (80), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 10/89 (11%)
Query 172 QRQAEAYRPLED------PRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVR 225
+RQ E R E R QG L+V L +++LR+EF +G + +V+
Sbjct 271 ERQTELKRKFEQMKQDRMTRYQG---VNLYVKNLDDGLDDERLRKEFSPFGTITSAKVMM 327
Query 226 DPRGRPRGYAFVEFERDRDMKEAYKKADG 254
+ GR +G+ FV F + +A + +G
Sbjct 328 E-GGRSKGFGFVCFSSPEEATKAVTEMNG 355
> hsa:5940 RBMY1A1, MGC181956, RBM1, RBM2, RBMY, RBMY1C, YRRM1,
YRRM2; RNA binding motif protein, Y-linked, family 1, member
A1
Length=496
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 47/80 (58%), Gaps = 0/80 (0%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDM 245
++ D LF+G L+ ET+EK L+ F +GP+ V +++D + RG+AF+ FE D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 246 KEAYKKADGTKIDGRRVLVD 265
K A K +G + G+ + V+
Sbjct 62 KNAAKDMNGKSLHGKAIKVE 81
> hsa:378950 RBMY1E; RNA binding motif protein, Y-linked, family
1, member E
Length=496
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 47/80 (58%), Gaps = 0/80 (0%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDM 245
++ D LF+G L+ ET+EK L+ F +GP+ V +++D + RG+AF+ FE D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 246 KEAYKKADGTKIDGRRVLVD 265
K A K +G + G+ + V+
Sbjct 62 KNAAKDMNGKSLHGKAIKVE 81
> hsa:378949 RBMY1D; RNA binding motif protein, Y-linked, family
1, member D
Length=496
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 47/80 (58%), Gaps = 0/80 (0%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDM 245
++ D LF+G L+ ET+EK L+ F +GP+ V +++D + RG+AF+ FE D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 246 KEAYKKADGTKIDGRRVLVD 265
K A K +G + G+ + V+
Sbjct 62 KNAAKDMNGKSLHGKAIKVE 81
> xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) binding
protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=626
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 40/74 (54%), Gaps = 0/74 (0%)
Query 191 FATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDMKEAYK 250
F +++ + +++L+ F YG V+V+ DP G+ +G+ FV FER D +A
Sbjct 190 FTNVYIKNFGEDMDDERLKETFSKYGKTLSVKVMTDPSGKSKGFGFVSFERHEDANKAVD 249
Query 251 KADGTKIDGRRVLV 264
+G ++G+ + V
Sbjct 250 DMNGKDVNGKIMFV 263
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 1/82 (1%)
Query 183 DPRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERD 242
DP ++ +F+ L K L F +G + +VV D G +GYAFV FE
Sbjct 90 DPSLRKSGVGNVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGS-KGYAFVHFETQ 148
Query 243 RDMKEAYKKADGTKIDGRRVLV 264
A +K +G ++ R+V V
Sbjct 149 DAADRAIEKMNGMLLNDRKVFV 170
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 1/74 (1%)
Query 190 PFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRG-RPRGYAFVEFERDRDMKEA 248
P A+L+VG L + +E L +F GPV +RV RD R GYA+V F++ D + A
Sbjct 9 PMASLYVGDLHPDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERA 68
Query 249 YKKADGTKIDGRRV 262
+ I G+ +
Sbjct 69 LDTMNFDVIKGKPI 82
Score = 38.9 bits (89), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 47/99 (47%), Gaps = 10/99 (10%)
Query 172 QRQAEAYRPLED------PRIQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVR 225
+RQAE R E R QG L++ L ++KLR+EF +G + +V+
Sbjct 271 ERQAELKRRFEQLKQERISRYQG---VNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVML 327
Query 226 DPRGRPRGYAFVEFERDRDMKEAYKKADGTKIDGRRVLV 264
+ GR +G+ FV F + +A + +G + + + V
Sbjct 328 E-EGRSKGFGFVCFSSPEEATKAVTEMNGRIVGSKPLYV 365
> hsa:378948 RBMY1B; RNA binding motif protein, Y-linked, family
1, member B
Length=496
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 47/80 (58%), Gaps = 0/80 (0%)
Query 186 IQGDPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDPRGRPRGYAFVEFERDRDM 245
++ D LF+G L+ ET+EK L+ F +GP+ V +++D + RG+AF+ FE D
Sbjct 2 VEADHPGKLFIGGLNRETNEKMLKAVFGKHGPISEVLLIKDRTSKSRGFAFITFENPADA 61
Query 246 KEAYKKADGTKIDGRRVLVD 265
K A K +G + G+ + V+
Sbjct 62 KNAAKDMNGKSLHGKAIKVE 81
> ath:AT4G39260 GR-RBP8; RNA binding / nucleic acid binding /
nucleotide binding
Length=169
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 48/72 (66%), Gaps = 1/72 (1%)
Query 195 FVGKLSYETSEKKLRREFEVYGPVRRVRVVRD-PRGRPRGYAFVEFERDRDMKEAYKKAD 253
FVG L++ T+++ L+R F +G V +++ D GR RG+ FV F+ ++ M++A ++ +
Sbjct 9 FVGGLAWATNDEDLQRTFSQFGDVIDSKIINDRESGRSRGFGFVTFKDEKAMRDAIEEMN 68
Query 254 GTKIDGRRVLVD 265
G ++DGR + V+
Sbjct 69 GKELDGRVITVN 80
> ath:AT3G26420 ATRZ-1A; RNA binding / nucleotide binding
Length=245
Score = 57.8 bits (138), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 45/78 (57%), Gaps = 1/78 (1%)
Query 189 DPFATLFVGKLSYETSEKKLRREFEVYGPVRRVRVVRDP-RGRPRGYAFVEFERDRDMKE 247
DP F+G L++ TS++ LR FE YG + +VV D GR RG+ F+ F+ + M E
Sbjct 4 DPEYRCFIGGLAWTTSDRGLRDAFEKYGHLVEAKVVLDKFSGRSRGFGFITFDEKKAMDE 63
Query 248 AYKKADGTKIDGRRVLVD 265
A +G +DGR + VD
Sbjct 64 AIAAMNGMDLDGRTITVD 81
Lambda K H
0.324 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10848096440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40