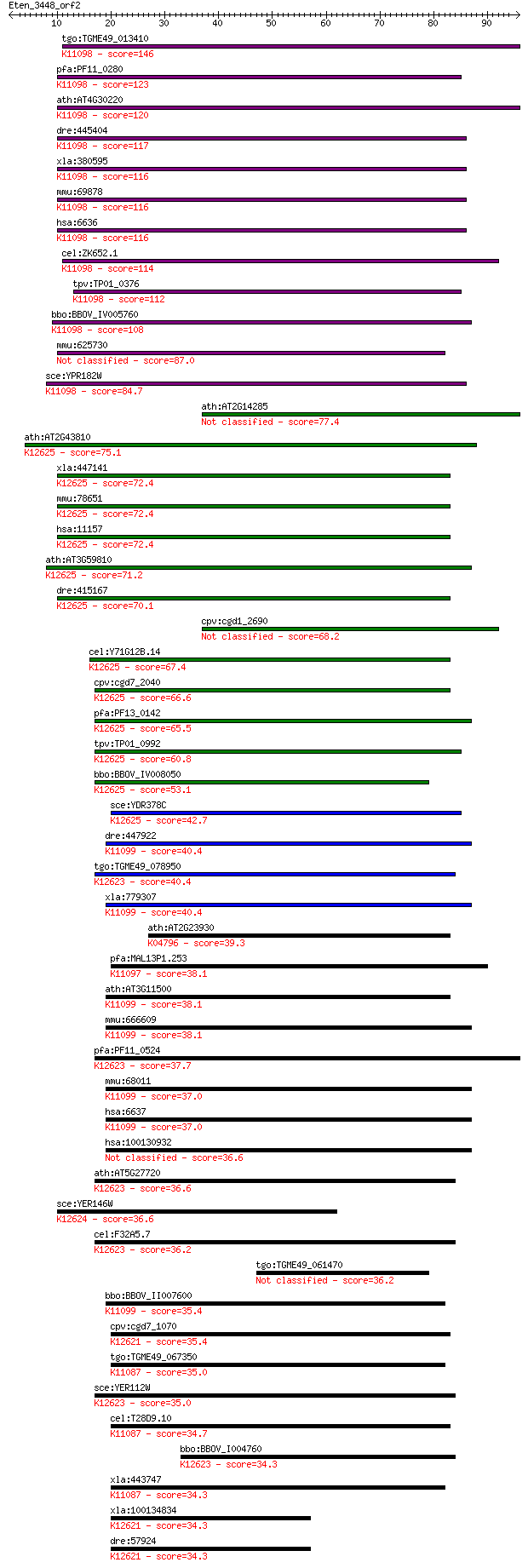
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3448_orf2
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_013410 small nuclear ribonucleoprotein F, putative ... 146 2e-35
pfa:PF11_0280 small nuclear ribonucleoprotein F, putative; K11... 123 2e-28
ath:AT4G30220 RUXF; RUXF (SMALL NUCLEAR RIBONUCLEOPROTEIN F); ... 120 1e-27
dre:445404 snrpf, snrpfl; small nuclear ribonucleoprotein poly... 117 9e-27
xla:380595 snrpf, MGC69162; small nuclear ribonucleoprotein po... 116 1e-26
mmu:69878 Snrpf, 2010013O18Rik, SMF; small nuclear ribonucleop... 116 1e-26
hsa:6636 SNRPF, SMF, Sm-F; small nuclear ribonucleoprotein pol... 116 1e-26
cel:ZK652.1 snr-5; Small Nuclear Ribonucleoprotein family memb... 114 1e-25
tpv:TP01_0376 small nuclear ribonucleoprotein F; K11098 small ... 112 3e-25
bbo:BBOV_IV005760 23.m05936; small nuclear ribonucleoprotein F... 108 5e-24
mmu:625730 Gm6616, EG625730; predicted gene 6616 87.0
sce:YPR182W SMX3; Sm F; SmF; K11098 small nuclear ribonucleopr... 84.7 7e-17
ath:AT2G14285 hypothetical protein 77.4 1e-14
ath:AT2G43810 small nuclear ribonucleoprotein F, putative / U6... 75.1 5e-14
xla:447141 lsm6; LSM6 homolog, U6 small nuclear RNA associated... 72.4 3e-13
mmu:78651 Lsm6, 1500031N17Rik, 2410088K19Rik, AI747288, MGC117... 72.4 3e-13
hsa:11157 LSM6, YDR378C; LSM6 homolog, U6 small nuclear RNA as... 72.4 3e-13
ath:AT3G59810 small nuclear ribonucleoprotein F, putative / U6... 71.2 8e-13
dre:415167 lsm6, zgc:92379; LSM6 homolog, U6 small nuclear RNA... 70.1 2e-12
cpv:cgd1_2690 hypothetical protein 68.2 7e-12
cel:Y71G12B.14 lsm-6; LSM Sm-like protein family member (lsm-6... 67.4 1e-11
cpv:cgd7_2040 small nuclear ribonucleo protein ; K12625 U6 snR... 66.6 2e-11
pfa:PF13_0142 Lsm6 homologue, putative; K12625 U6 snRNA-associ... 65.5 4e-11
tpv:TP01_0992 U6 small nuclear ribonucleoprotein; K12625 U6 sn... 60.8 1e-09
bbo:BBOV_IV008050 23.m06295; small nuclear ribonucleoprotein F... 53.1 2e-07
sce:YDR378C LSM6; Lsm (Like Sm) protein; part of heteroheptame... 42.7 3e-04
dre:447922 zgc:103688; K11099 small nuclear ribonucleoprotein G 40.4 0.001
tgo:TGME49_078950 U6 snRNA associated Sm-like protein LSm4, pu... 40.4 0.001
xla:779307 snrpg, MGC154809; small nuclear ribonucleoprotein p... 40.4 0.002
ath:AT2G23930 SNRNP-G (PROBABLE SMALL NUCLEAR RIBONUCLEOPROTEI... 39.3 0.003
pfa:MAL13P1.253 small nuclear ribonucleoprotein, putative; K11... 38.1 0.006
ath:AT3G11500 small nuclear ribonucleoprotein G, putative / sn... 38.1 0.007
mmu:666609 Gm8186, EG666609; predicted gene 8186; K11099 small... 38.1 0.007
pfa:PF11_0524 lsm4 homologue, putative; K12623 U6 snRNA-associ... 37.7 0.008
mmu:68011 Snrpg, 2810024K17Rik, AL022803, SMG; small nuclear r... 37.0 0.014
hsa:6637 SNRPG, MGC117317, SMG, Sm-G; small nuclear ribonucleo... 37.0 0.014
hsa:100130932 small nuclear ribonucleoprotein G-like protein 36.6 0.018
ath:AT5G27720 emb1644 (embryo defective 1644); K12623 U6 snRNA... 36.6 0.019
sce:YER146W LSM5; Lsm (Like Sm) protein; part of heteroheptame... 36.6 0.021
cel:F32A5.7 lsm-4; LSM Sm-like protein family member (lsm-4); ... 36.2 0.024
tgo:TGME49_061470 U6 snRNA-associated Sm-like protein, putative 36.2 0.025
bbo:BBOV_II007600 18.m06632; small nuclear ribonucleoprotein G... 35.4 0.042
cpv:cgd7_1070 snRNP core protein ; K12621 U6 snRNA-associated ... 35.4 0.046
tgo:TGME49_067350 small nuclear ribonucleoprotein, putative ; ... 35.0 0.054
sce:YER112W LSM4, SDB23, USS1; Lsm (Like Sm) protein; part of ... 35.0 0.067
cel:T28D9.10 snr-3; Small Nuclear Ribonucleoprotein family mem... 34.7 0.078
bbo:BBOV_I004760 19.m02880; U6 snRNA-associated Sm-like protei... 34.3 0.090
xla:443747 snrpd1, MGC130717, MGC160444, MGC78968; small nucle... 34.3 0.093
xla:100134834 lsm2, MGC84990; LSM2 homolog, U6 small nuclear R... 34.3 0.095
dre:57924 smx5, lsm2, wu:fe48h10, zgc:101795; smx5; K12621 U6 ... 34.3 0.096
> tgo:TGME49_013410 small nuclear ribonucleoprotein F, putative
; K11098 small nuclear ribonucleoprotein F
Length=87
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 64/85 (75%), Positives = 79/85 (92%), Gaps = 0/85 (0%)
Query 11 SLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLG 70
++ P+NPKPFLTSLTG+QVMVKLKWG+EYKGYLKSFD+YMN+ L N+EEWV+G+FKG LG
Sbjct 3 TVTPVNPKPFLTSLTGRQVMVKLKWGIEYKGYLKSFDEYMNLQLQNTEEWVDGSFKGHLG 62
Query 71 EVLIRCNNVLYIRQVQGEDEAEEAS 95
EVL+RCNNVLY+RQV+GEDE +E +
Sbjct 63 EVLLRCNNVLYLRQVKGEDEEDEEA 87
> pfa:PF11_0280 small nuclear ribonucleoprotein F, putative; K11098
small nuclear ribonucleoprotein F
Length=86
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 52/75 (69%), Positives = 64/75 (85%), Gaps = 0/75 (0%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
+ +APLNPKPFL SL G +V++KLKWGMEYKG LKSFD YMNI L N+EEW+ G FKG+L
Sbjct 3 VGIAPLNPKPFLNSLAGNRVIIKLKWGMEYKGILKSFDGYMNIRLTNAEEWIHGEFKGTL 62
Query 70 GEVLIRCNNVLYIRQ 84
GE+ +RCNN+LYIR+
Sbjct 63 GEIFLRCNNILYIRE 77
> ath:AT4G30220 RUXF; RUXF (SMALL NUCLEAR RIBONUCLEOPROTEIN F);
K11098 small nuclear ribonucleoprotein F
Length=88
Score = 120 bits (301), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 53/86 (61%), Positives = 69/86 (80%), Gaps = 0/86 (0%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
M+ P+NPKPFL +LTGK V+VKLKWGMEYKG+L S D YMN+ L N+EE+++G G+L
Sbjct 1 MATIPVNPKPFLNNLTGKTVIVKLKWGMEYKGFLASVDSYMNLQLGNTEEYIDGQLTGNL 60
Query 70 GEVLIRCNNVLYIRQVQGEDEAEEAS 95
GE+LIRCNNVLY+R V ++E E+A
Sbjct 61 GEILIRCNNVLYVRGVPEDEELEDAD 86
> dre:445404 snrpf, snrpfl; small nuclear ribonucleoprotein polypeptide
F; K11098 small nuclear ribonucleoprotein F
Length=86
Score = 117 bits (293), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 58/76 (76%), Positives = 62/76 (81%), Gaps = 1/76 (1%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL PLNPKPFL LTGK VMVKLKWGMEYKGYL S D YMN+ L N+EE+V+G G L
Sbjct 1 MSL-PLNPKPFLNGLTGKPVMVKLKWGMEYKGYLVSVDGYMNMQLANTEEYVDGALAGHL 59
Query 70 GEVLIRCNNVLYIRQV 85
GEVLIRCNNVLYIR V
Sbjct 60 GEVLIRCNNVLYIRGV 75
> xla:380595 snrpf, MGC69162; small nuclear ribonucleoprotein
polypeptide F; K11098 small nuclear ribonucleoprotein F
Length=86
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 57/76 (75%), Positives = 62/76 (81%), Gaps = 1/76 (1%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL PLNPKPFL LTGK VMVKLKWGMEYKGYL S D YMN+ L N+EE+++G G L
Sbjct 1 MSL-PLNPKPFLNGLTGKPVMVKLKWGMEYKGYLVSVDGYMNMQLANTEEYIDGALSGHL 59
Query 70 GEVLIRCNNVLYIRQV 85
GEVLIRCNNVLYIR V
Sbjct 60 GEVLIRCNNVLYIRGV 75
> mmu:69878 Snrpf, 2010013O18Rik, SMF; small nuclear ribonucleoprotein
polypeptide F; K11098 small nuclear ribonucleoprotein
F
Length=86
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 57/76 (75%), Positives = 62/76 (81%), Gaps = 1/76 (1%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL PLNPKPFL LTGK VMVKLKWGMEYKGYL S D YMN+ L N+EE+++G G L
Sbjct 1 MSL-PLNPKPFLNGLTGKPVMVKLKWGMEYKGYLVSVDGYMNMQLANTEEYIDGALSGHL 59
Query 70 GEVLIRCNNVLYIRQV 85
GEVLIRCNNVLYIR V
Sbjct 60 GEVLIRCNNVLYIRGV 75
> hsa:6636 SNRPF, SMF, Sm-F; small nuclear ribonucleoprotein polypeptide
F; K11098 small nuclear ribonucleoprotein F
Length=86
Score = 116 bits (291), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 57/76 (75%), Positives = 62/76 (81%), Gaps = 1/76 (1%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL PLNPKPFL LTGK VMVKLKWGMEYKGYL S D YMN+ L N+EE+++G G L
Sbjct 1 MSL-PLNPKPFLNGLTGKPVMVKLKWGMEYKGYLVSVDGYMNMQLANTEEYIDGALSGHL 59
Query 70 GEVLIRCNNVLYIRQV 85
GEVLIRCNNVLYIR V
Sbjct 60 GEVLIRCNNVLYIRGV 75
> cel:ZK652.1 snr-5; Small Nuclear Ribonucleoprotein family member
(snr-5); K11098 small nuclear ribonucleoprotein F
Length=85
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 50/81 (61%), Positives = 66/81 (81%), Gaps = 0/81 (0%)
Query 11 SLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLG 70
++ P+NPKPFL SLTGK V+ KLKWGMEYKG L + D YMN+ L ++EE+++GN +G+LG
Sbjct 3 AVQPVNPKPFLNSLTGKFVVCKLKWGMEYKGVLVAVDSYMNLQLAHAEEYIDGNSQGNLG 62
Query 71 EVLIRCNNVLYIRQVQGEDEA 91
E+LIRCNNVLY+ V GE+E
Sbjct 63 EILIRCNNVLYVGGVDGENET 83
> tpv:TP01_0376 small nuclear ribonucleoprotein F; K11098 small
nuclear ribonucleoprotein F
Length=91
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 52/73 (71%), Positives = 60/73 (82%), Gaps = 1/73 (1%)
Query 13 APLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLG-E 71
APLNPKPFLTSL GK V+VKLKWGMEYKG LKSFD YMN+ L + EEW G KG LG +
Sbjct 12 APLNPKPFLTSLAGKTVVVKLKWGMEYKGTLKSFDSYMNLQLSDPEEWENGVLKGKLGND 71
Query 72 VLIRCNNVLYIRQ 84
+LIRCNN+LY+R+
Sbjct 72 ILIRCNNMLYVRE 84
> bbo:BBOV_IV005760 23.m05936; small nuclear ribonucleoprotein
F; K11098 small nuclear ribonucleoprotein F
Length=94
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 50/79 (63%), Positives = 60/79 (75%), Gaps = 1/79 (1%)
Query 9 EMSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGS 68
+ ++ PLNPKPFL L G+ V V LKWGMEYKG+LKSFD YMNI L N+EEW G KG+
Sbjct 16 QQAVTPLNPKPFLAKLVGQNVFVTLKWGMEYKGFLKSFDSYMNIELENAEEWENGVMKGT 75
Query 69 L-GEVLIRCNNVLYIRQVQ 86
L G +LIRCNNVL+IR +
Sbjct 76 LGGTILIRCNNVLHIRAAE 94
> mmu:625730 Gm6616, EG625730; predicted gene 6616
Length=83
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/72 (65%), Positives = 52/72 (72%), Gaps = 4/72 (5%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL PLNPK L LTGK VMVKLKWGMEY L S D YMN+ L N+EE+++G L
Sbjct 1 MSL-PLNPKTSLNGLTGKPVMVKLKWGMEY---LVSMDGYMNMQLANTEEYIDGELSEHL 56
Query 70 GEVLIRCNNVLY 81
GEVL RCNNVLY
Sbjct 57 GEVLKRCNNVLY 68
> sce:YPR182W SMX3; Sm F; SmF; K11098 small nuclear ribonucleoprotein
F
Length=86
Score = 84.7 bits (208), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 41/79 (51%), Positives = 55/79 (69%), Gaps = 1/79 (1%)
Query 8 DEMSLAPLNPKPFLTSLTGKQVMVKLKW-GMEYKGYLKSFDDYMNIHLLNSEEWVEGNFK 66
D ++ P+NPKPFL L +V VKLK+ EY+G L S D+Y N+ L +EE+V G
Sbjct 6 DISAMQPVNPKPFLKGLVNHRVGVKLKFNSTEYRGTLVSTDNYFNLQLNEAEEFVAGVSH 65
Query 67 GSLGEVLIRCNNVLYIRQV 85
G+LGE+ IRCNNVLYIR++
Sbjct 66 GTLGEIFIRCNNVLYIREL 84
> ath:AT2G14285 hypothetical protein
Length=61
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/59 (57%), Positives = 46/59 (77%), Gaps = 0/59 (0%)
Query 37 MEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNNVLYIRQVQGEDEAEEAS 95
MEYKG+L S D YMN+ L N+EE+++G G+LGE+LIRCNNVLY+R V ++E E+A
Sbjct 1 MEYKGFLASVDSYMNLQLGNTEEYIDGQLTGNLGEILIRCNNVLYVRGVPEDEELEDAD 59
> ath:AT2G43810 small nuclear ribonucleoprotein F, putative /
U6 snRNA-associated Sm-like protein, putative / Sm protein F,
putative; K12625 U6 snRNA-associated Sm-like protein LSm6
Length=91
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 50/84 (59%), Gaps = 0/84 (0%)
Query 4 TSFGDEMSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEG 63
+ G++ S P FL S+ GK V+VKL G++Y+G L D YMNI + +EE+V G
Sbjct 2 SGVGEKASGTTKTPADFLKSIRGKPVVVKLNSGVDYRGILTCLDGYMNIAMEQTEEYVNG 61
Query 64 NFKGSLGEVLIRCNNVLYIRQVQG 87
K + G+ +R NNVLYI +G
Sbjct 62 QLKNTYGDAFVRGNNVLYISTTKG 85
> xla:447141 lsm6; LSM6 homolog, U6 small nuclear RNA associated;
K12625 U6 snRNA-associated Sm-like protein LSm6
Length=80
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL P FL + G+ V+VKL G++Y+G L D YMNI L +EE+V G K
Sbjct 1 MSLRKQTPSDFLKQIIGRPVVVKLNSGVDYRGVLACLDGYMNIALEQTEEYVNGQLKNKY 60
Query 70 GEVLIRCNNVLYI 82
G+ IR NNVLYI
Sbjct 61 GDAFIRGNNVLYI 73
> mmu:78651 Lsm6, 1500031N17Rik, 2410088K19Rik, AI747288, MGC117708;
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae);
K12625 U6 snRNA-associated Sm-like protein LSm6
Length=80
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL P FL + G+ V+VKL G++Y+G L D YMNI L +EE+V G K
Sbjct 1 MSLRKQTPSDFLKQIIGRPVVVKLNSGVDYRGVLACLDGYMNIALEQTEEYVNGQLKNKY 60
Query 70 GEVLIRCNNVLYI 82
G+ IR NNVLYI
Sbjct 61 GDAFIRGNNVLYI 73
> hsa:11157 LSM6, YDR378C; LSM6 homolog, U6 small nuclear RNA
associated (S. cerevisiae); K12625 U6 snRNA-associated Sm-like
protein LSm6
Length=80
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/73 (47%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL P FL + G+ V+VKL G++Y+G L D YMNI L +EE+V G K
Sbjct 1 MSLRKQTPSDFLKQIIGRPVVVKLNSGVDYRGVLACLDGYMNIALEQTEEYVNGQLKNKY 60
Query 70 GEVLIRCNNVLYI 82
G+ IR NNVLYI
Sbjct 61 GDAFIRGNNVLYI 73
> ath:AT3G59810 small nuclear ribonucleoprotein F, putative /
U6 snRNA-associated Sm-like protein, putative / Sm protein F,
putative; K12625 U6 snRNA-associated Sm-like protein LSm6
Length=91
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 34/79 (43%), Positives = 47/79 (59%), Gaps = 0/79 (0%)
Query 8 DEMSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKG 67
+++S P FL S+ G+ V+VKL G++Y+G L D YMNI + +EE+V G K
Sbjct 6 EKVSGTTKTPADFLKSIRGRPVVVKLNSGVDYRGTLTCLDGYMNIAMEQTEEYVNGQLKN 65
Query 68 SLGEVLIRCNNVLYIRQVQ 86
G+ IR NNVLYI V
Sbjct 66 KYGDAFIRGNNVLYISTVN 84
> dre:415167 lsm6, zgc:92379; LSM6 homolog, U6 small nuclear RNA
associated (S. cerevisiae); K12625 U6 snRNA-associated Sm-like
protein LSm6
Length=80
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSL 69
MSL P FL + G+ V+VKL G++Y+G L D YMNI + +EE+V G K
Sbjct 1 MSLRKQTPSDFLKQIIGRPVVVKLNSGVDYRGVLACLDGYMNIAVEQTEEYVNGQLKNKY 60
Query 70 GEVLIRCNNVLYI 82
G+ +R NNVLYI
Sbjct 61 GDAFLRGNNVLYI 73
> cpv:cgd1_2690 hypothetical protein
Length=67
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 37 MEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNNVLYIRQVQGEDEA 91
M YKG L SFD+YMNI L + EEW++ KG+LG+V IRCNNVLYI + + E
Sbjct 1 MRYKGTLVSFDEYMNILLNDCEEWIDNTKKGTLGKVFIRCNNVLYISRAEESKET 55
> cel:Y71G12B.14 lsm-6; LSM Sm-like protein family member (lsm-6);
K12625 U6 snRNA-associated Sm-like protein LSm6
Length=77
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 32/67 (47%), Positives = 41/67 (61%), Gaps = 0/67 (0%)
Query 16 NPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIR 75
NP FL + GK V+VKL G++Y+G L D YMNI L +EE+ G + G+ IR
Sbjct 6 NPAEFLKKVIGKPVVVKLNSGVDYRGILACLDGYMNIALEQTEEYSNGQLQNKYGDAFIR 65
Query 76 CNNVLYI 82
NNVLYI
Sbjct 66 GNNVLYI 72
> cpv:cgd7_2040 small nuclear ribonucleo protein ; K12625 U6 snRNA-associated
Sm-like protein LSm6
Length=93
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 43/66 (65%), Gaps = 0/66 (0%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRC 76
P FL + G V+V+L G++YKG L DD MNI + ++EE+V+G F + G+ IR
Sbjct 23 PSDFLRQVIGSPVIVRLNSGVDYKGLLACLDDRMNIAMEDTEEYVDGVFVENHGDTFIRG 82
Query 77 NNVLYI 82
NNVLYI
Sbjct 83 NNVLYI 88
> pfa:PF13_0142 Lsm6 homologue, putative; K12625 U6 snRNA-associated
Sm-like protein LSm6
Length=77
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRC 76
PK F+ SL G+ V+V+L G +YKG L D+ MN+ L +EE+ +G + IR
Sbjct 6 PKDFVESLKGRAVIVRLNNGTDYKGILACLDERMNVALEQTEEYYDGELTDKYNDAFIRG 65
Query 77 NNVLYIRQVQ 86
NNV YIR ++
Sbjct 66 NNVFYIRAIE 75
> tpv:TP01_0992 U6 small nuclear ribonucleoprotein; K12625 U6
snRNA-associated Sm-like protein LSm6
Length=77
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 43/68 (63%), Gaps = 0/68 (0%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRC 76
P ++TS+T K V+VKL G ++KG L S D+ MN+ + N+ E+++G S G+ IR
Sbjct 6 PSAYITSITRKPVLVKLNNGSDFKGTLASLDERMNLAMENTCEFLDGVMVKSYGDAFIRG 65
Query 77 NNVLYIRQ 84
NNV YI
Sbjct 66 NNVYYISH 73
> bbo:BBOV_IV008050 23.m06295; small nuclear ribonucleoprotein
F; K12625 U6 snRNA-associated Sm-like protein LSm6
Length=83
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRC 76
P ++ ++T K V+VKL G ++G L S DD MN+ + N++E+ +G + G+ IR
Sbjct 6 PSSYIANITRKPVIVKLNNGTRFRGLLSSLDDRMNLAMENTKEYCDGELVKAYGDSFIRG 65
Query 77 NN 78
NN
Sbjct 66 NN 67
> sce:YDR378C LSM6; Lsm (Like Sm) protein; part of heteroheptameric
complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic
Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex
part of U6 snRNP and possibly involved in processing tRNA,
snoRNA, and rRNA; K12625 U6 snRNA-associated Sm-like protein
LSm6
Length=86
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 36/69 (52%), Gaps = 4/69 (5%)
Query 20 FLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEG----NFKGSLGEVLIR 75
FL+ + GK V VKL G+ Y G L+S D +MN+ L ++ E E +V +R
Sbjct 15 FLSDIIGKTVNVKLASGLLYSGRLESIDGFMNVALSSATEHYESNNNKLLNKFNSDVFLR 74
Query 76 CNNVLYIRQ 84
V+YI +
Sbjct 75 GTQVMYISE 83
> dre:447922 zgc:103688; K11099 small nuclear ribonucleoprotein
G
Length=76
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 38/68 (55%), Gaps = 0/68 (0%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L K++ +KL G +G L+ FD +MN+ + ++ E G ++G V+IR N+
Sbjct 7 PELKKFMDKKLSLKLNGGRHVQGILRGFDPFMNLVVDDTIEMAPGGQMNTIGMVVIRGNS 66
Query 79 VLYIRQVQ 86
++ + ++
Sbjct 67 IIMLEALE 74
> tgo:TGME49_078950 U6 snRNA associated Sm-like protein LSm4,
putative ; K12623 U6 snRNA-associated Sm-like protein LSm4
Length=214
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSE-EWVEGNFKGSLGEVLIR 75
P L + + +MV+LK G Y G L S D +MN+HL +S +G L E IR
Sbjct 4 PLTLLRAAQNRPMMVELKSGETYSGLLASCDGFMNLHLKDSVCTSKDGERFWKLSECYIR 63
Query 76 CNNVLYIR 83
N V YIR
Sbjct 64 GNMVKYIR 71
> xla:779307 snrpg, MGC154809; small nuclear ribonucleoprotein
polypeptide G; K11099 small nuclear ribonucleoprotein G
Length=76
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 38/68 (55%), Gaps = 0/68 (0%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L K++ +K+ G + +G L+ FD +MN+ L S E + S+G V+IR N+
Sbjct 7 PELKKFMDKKLSLKINGGRQVQGILRGFDPFMNLVLDESTEISGSGNQNSIGMVVIRGNS 66
Query 79 VLYIRQVQ 86
++ + ++
Sbjct 67 IIMLEALE 74
> ath:AT2G23930 SNRNP-G (PROBABLE SMALL NUCLEAR RIBONUCLEOPROTEIN
G); K04796 small nuclear ribonucleoprotein
Length=67
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Query 27 KQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNNVLYI 82
K++ +KL G L+ FD +MN+ + N+ E V GN K +G V+IR N+++ +
Sbjct 3 KKLQIKLNANRMVTGTLRGFDQFMNLVVDNTVE-VNGNDKTDIGMVVIRGNSIVTV 57
> pfa:MAL13P1.253 small nuclear ribonucleoprotein, putative; K11097
small nuclear ribonucleoprotein E
Length=93
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 42/71 (59%), Gaps = 1/71 (1%)
Query 20 FLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEW-VEGNFKGSLGEVLIRCNN 78
F T+ T Q+ + K M +G + FD+YMN+ L ++E V+ N K LG++L++ +
Sbjct 22 FFTNKTVVQIWLYDKPDMRIEGIILGFDEYMNMVLDQTKEISVKKNTKKELGKILLKGDT 81
Query 79 VLYIRQVQGED 89
+ I +V+ E+
Sbjct 82 ITLIMEVKNEE 92
> ath:AT3G11500 small nuclear ribonucleoprotein G, putative /
snRNP-G, putative / Sm protein G, putative; K11099 small nuclear
ribonucleoprotein G
Length=79
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 36/64 (56%), Gaps = 1/64 (1%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L K++ +KL G L+ FD +MN+ + N+ E V G+ K +G V+IR N+
Sbjct 8 PDLKKYMDKKLQIKLNANRMVVGTLRGFDQFMNLVVDNTVE-VNGDDKTDIGMVVIRGNS 66
Query 79 VLYI 82
++ +
Sbjct 67 IVTV 70
> mmu:666609 Gm8186, EG666609; predicted gene 8186; K11099 small
nuclear ribonucleoprotein G
Length=76
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 38/68 (55%), Gaps = 0/68 (0%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L + G+++ +KL G +G L+ FD +MN+ + E + ++G V+IR N+
Sbjct 7 PELKKVYGQELSLKLNGGRHVQGILQGFDPFMNLVIDECVEMATSGQQNNIGMVVIRGNS 66
Query 79 VLYIRQVQ 86
++ + ++
Sbjct 67 IIMLEALE 74
> pfa:PF11_0524 lsm4 homologue, putative; K12623 U6 snRNA-associated
Sm-like protein LSm4
Length=126
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 42/81 (51%), Gaps = 2/81 (2%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLN-SEEWVEGNFKGSLGEVLIR 75
P L + VMV+LK G Y G+L D +MN+H+ N +G+ + E +R
Sbjct 4 PLTLLKCSQNQPVMVELKNGETYSGFLVFCDRFMNLHMKNIICTSKDGDKFWKISECYVR 63
Query 76 CNNVLYIR-QVQGEDEAEEAS 95
N++ YIR Q Q ++A E +
Sbjct 64 GNSIKYIRVQDQAIEQAIEET 84
> mmu:68011 Snrpg, 2810024K17Rik, AL022803, SMG; small nuclear
ribonucleoprotein polypeptide G; K11099 small nuclear ribonucleoprotein
G
Length=76
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 36/68 (52%), Gaps = 0/68 (0%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L K++ +KL G +G L+ FD +MN+ + E + ++G V+IR N+
Sbjct 7 PELKKFMDKKLSLKLNGGRHVQGILRGFDPFMNLVIDECVEMATSGQQNNIGMVVIRGNS 66
Query 79 VLYIRQVQ 86
++ + ++
Sbjct 67 IIMLEALE 74
> hsa:6637 SNRPG, MGC117317, SMG, Sm-G; small nuclear ribonucleoprotein
polypeptide G; K11099 small nuclear ribonucleoprotein
G
Length=76
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 36/68 (52%), Gaps = 0/68 (0%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L K++ +KL G +G L+ FD +MN+ + E + ++G V+IR N+
Sbjct 7 PELKKFMDKKLSLKLNGGRHVQGILRGFDPFMNLVIDECVEMATSGQQNNIGMVVIRGNS 66
Query 79 VLYIRQVQ 86
++ + ++
Sbjct 67 IIMLEALE 74
> hsa:100130932 small nuclear ribonucleoprotein G-like protein
Length=74
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 35/68 (51%), Gaps = 0/68 (0%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L T K+ +KL G +G L+ FD +MN+ + E + ++G V IR N+
Sbjct 7 PELKKFTDKKFSLKLNGGRHVQGILRGFDPFMNLVIDECVEMATSGQQKNIGMVEIRGNS 66
Query 79 VLYIRQVQ 86
++ + ++
Sbjct 67 IIMLEALE 74
> ath:AT5G27720 emb1644 (embryo defective 1644); K12623 U6 snRNA-associated
Sm-like protein LSm4
Length=129
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 37/71 (52%), Gaps = 7/71 (9%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWV----EGNFKGSLGEV 72
P L + G ++V+LK G Y G+L + D +MNIHL E + +G+ + E
Sbjct 3 PLSLLKTAQGHPMLVELKNGETYNGHLVNCDTWMNIHL---REVICTSKDGDRFWRMPEC 59
Query 73 LIRCNNVLYIR 83
IR N + Y+R
Sbjct 60 YIRGNTIKYLR 70
> sce:YER146W LSM5; Lsm (Like Sm) protein; part of heteroheptameric
complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic
Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex
part of U6 snRNP and possibly involved in processing tRNA,
snoRNA, and rRNA; K12624 U6 snRNA-associated Sm-like protein
LSm5
Length=93
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 31/52 (59%), Gaps = 0/52 (0%)
Query 10 MSLAPLNPKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWV 61
MSL + P + ++V++ L+ E++G L FDD++N+ L ++ EW+
Sbjct 1 MSLPEILPLEVIDKTINQKVLIVLQSNREFEGTLVGFDDFVNVILEDAVEWL 52
> cel:F32A5.7 lsm-4; LSM Sm-like protein family member (lsm-4);
K12623 U6 snRNA-associated Sm-like protein LSm4
Length=123
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 37/68 (54%), Gaps = 1/68 (1%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSE-EWVEGNFKGSLGEVLIR 75
P L + ++V+LK G Y G+LK+ D +MNIHL++ +G+ + E +R
Sbjct 4 PLSLLKTAQNHPMLVELKNGETYNGHLKACDSWMNIHLVDVIFTSKDGDKFFKMSEAYVR 63
Query 76 CNNVLYIR 83
+ + Y+R
Sbjct 64 GSTIKYLR 71
> tgo:TGME49_061470 U6 snRNA-associated Sm-like protein, putative
Length=86
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 47 DDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
DD MNI + +EE+V+GNF + G LIR NN
Sbjct 41 DDRMNIAMDKAEEFVDGNFVAAYGLSLIRGNN 72
> bbo:BBOV_II007600 18.m06632; small nuclear ribonucleoprotein
G; K11099 small nuclear ribonucleoprotein G
Length=80
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 1/63 (1%)
Query 19 PFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNN 78
P L K++ V L G L+ +D +MN+ L N+ + +EG+ + LG V+IR N
Sbjct 9 PDLRRYMEKRLDVHLTGSRHVVGVLRGYDTFMNVVLDNALQ-IEGDEQTRLGMVVIRGNA 67
Query 79 VLY 81
++Y
Sbjct 68 IVY 70
> cpv:cgd7_1070 snRNP core protein ; K12621 U6 snRNA-associated
Sm-like protein LSm2
Length=107
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 36/67 (53%), Gaps = 4/67 (5%)
Query 20 FLTSLTGKQVMV--KLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFK--GSLGEVLIR 75
FL +L KQV+V +LK ++ G L S D Y+NI L N+ +F GSL IR
Sbjct 10 FLQTLIEKQVVVTVELKNDLQITGTLHSIDQYLNIKLNNTTVNQNESFIHLGSLKNCFIR 69
Query 76 CNNVLYI 82
+ V YI
Sbjct 70 GSVVRYI 76
> tgo:TGME49_067350 small nuclear ribonucleoprotein, putative
; K11087 small nuclear ribonucleoprotein D1
Length=121
Score = 35.0 bits (79), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 20 FLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNNV 79
FL L + ++++LK G G + D MN HL N + ++ SL + IR NN+
Sbjct 6 FLMRLANETLVIELKNGTVIHGTVVGVDISMNTHLKNVKMTMKQGNPTSLEHLTIRGNNI 65
Query 80 LY 81
Y
Sbjct 66 RY 67
> sce:YER112W LSM4, SDB23, USS1; Lsm (Like Sm) protein; part of
heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p):
cytoplasmic Lsm1p complex involved in mRNA decay; nuclear
Lsm8p complex part of U6 snRNP and possibly involved in processing
tRNA, snoRNA, and rRNA; K12623 U6 snRNA-associated
Sm-like protein LSm4
Length=187
Score = 35.0 bits (79), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 40/78 (51%), Gaps = 11/78 (14%)
Query 17 PKPFLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHL-----------LNSEEWVEGNF 65
P LT+ G+Q+ ++LK G +G L + D++MN+ L +NSE+ E +
Sbjct 3 PLYLLTNAKGQQMQIELKNGEIIQGILTNVDNWMNLTLSNVTEYSEESAINSEDNAESSK 62
Query 66 KGSLGEVLIRCNNVLYIR 83
L E+ IR + +I+
Sbjct 63 AVKLNEIYIRGTFIKFIK 80
> cel:T28D9.10 snr-3; Small Nuclear Ribonucleoprotein family member
(snr-3); K11087 small nuclear ribonucleoprotein D1
Length=126
Score = 34.7 bits (78), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 0/63 (0%)
Query 20 FLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNNV 79
FL L+ + V ++LK G + G + D MN HL V+ L + IR NN+
Sbjct 6 FLMKLSHETVNIELKNGTQVSGTIMGVDVAMNTHLRAVSMTVKNKEPVKLDTLSIRGNNI 65
Query 80 LYI 82
YI
Sbjct 66 RYI 68
> bbo:BBOV_I004760 19.m02880; U6 snRNA-associated Sm-like protein
LSm4; K12623 U6 snRNA-associated Sm-like protein LSm4
Length=104
Score = 34.3 bits (77), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Query 33 LKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGS----LGEVLIRCNNVLYIR 83
+ +G Y G L S D +MN+H++N+ + + KG L E IR NNV R
Sbjct 8 ITFGETYSGILSSCDAFMNVHMINA---ICTSKKGDEFWKLSECFIRGNNVKSFR 59
> xla:443747 snrpd1, MGC130717, MGC160444, MGC78968; small nuclear
ribonucleoprotein D1 polypeptide 16kDa; K11087 small nuclear
ribonucleoprotein D1
Length=119
Score = 34.3 bits (77), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 30/62 (48%), Gaps = 0/62 (0%)
Query 20 FLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLNSEEWVEGNFKGSLGEVLIRCNNV 79
FL L+ + V V+LK G + G + D MN HL + V+ L + IR NN+
Sbjct 6 FLMKLSHETVTVELKNGTQVHGTITGVDVSMNTHLKAVKMTVKNREPVQLETLSIRGNNI 65
Query 80 LY 81
Y
Sbjct 66 RY 67
> xla:100134834 lsm2, MGC84990; LSM2 homolog, U6 small nuclear
RNA associated; K12621 U6 snRNA-associated Sm-like protein
LSm2
Length=95
Score = 34.3 bits (77), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 20 FLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLN 56
F SL GK V+V+LK + G L S D Y+NI L +
Sbjct 6 FFKSLVGKDVVVELKNDLSICGTLHSVDQYLNIKLTD 42
> dre:57924 smx5, lsm2, wu:fe48h10, zgc:101795; smx5; K12621 U6
snRNA-associated Sm-like protein LSm2
Length=95
Score = 34.3 bits (77), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 20 FLTSLTGKQVMVKLKWGMEYKGYLKSFDDYMNIHLLN 56
F SL GK V+V+LK + G L S D Y+NI L +
Sbjct 6 FFKSLVGKDVVVELKNDLSICGTLHSVDQYLNIKLTD 42
Lambda K H
0.315 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2065224908
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40