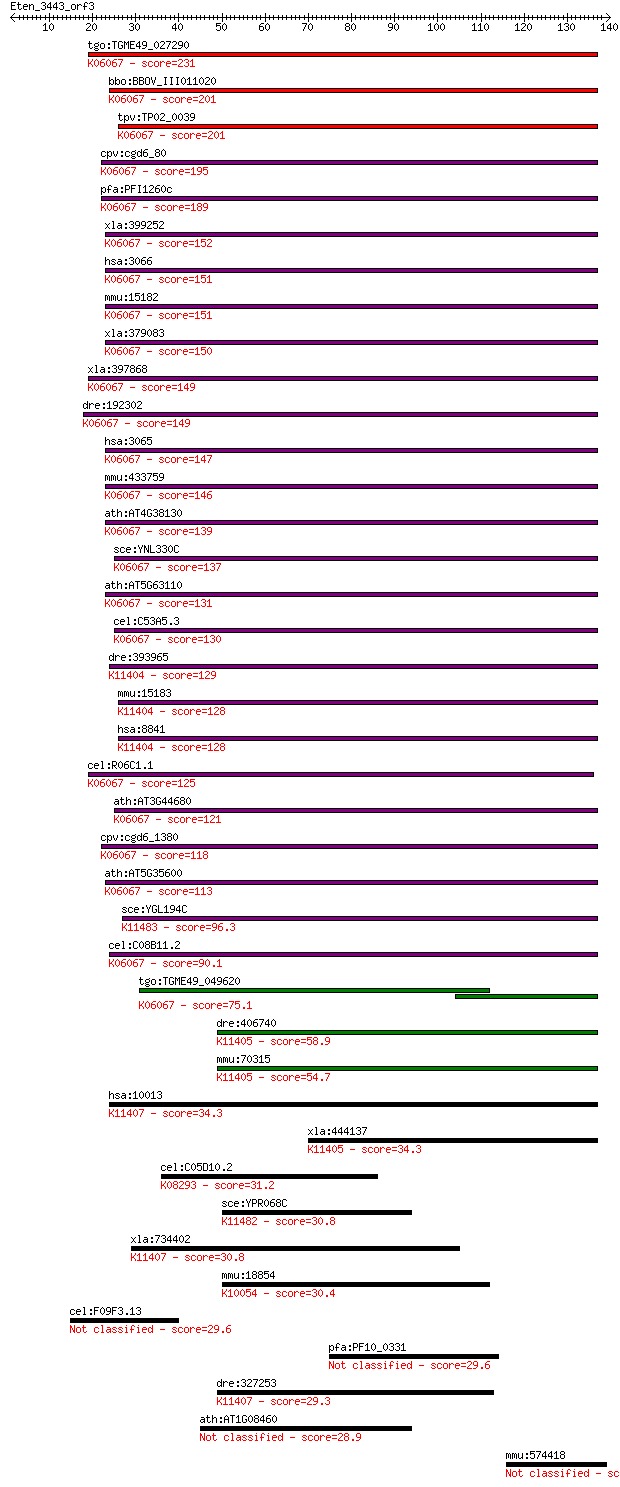
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3443_orf3
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_027290 histone deacetylase, putative ; K06067 histo... 231 6e-61
bbo:BBOV_III011020 17.m07949; histone deacetylase; K06067 hist... 201 4e-52
tpv:TP02_0039 histone deacetylase; K06067 histone deacetylase ... 201 8e-52
cpv:cgd6_80 RPD3/HD1 histone deacetylase ; K06067 histone deac... 195 4e-50
pfa:PFI1260c PfHDAC1; histone deacetylase; K06067 histone deac... 189 2e-48
xla:399252 hdac2, MGC81850; histone deacetylase 2 (EC:3.5.1.98... 152 2e-37
hsa:3066 HDAC2, HD2, RPD3, YAF1; histone deacetylase 2 (EC:3.5... 151 5e-37
mmu:15182 Hdac2, D10Wsu179e, YAF1, Yy1bp, mRPD3; histone deace... 151 6e-37
xla:379083 hdac1-b, HD1, MGC53583, RPD3, gon-10, hdac1, hdac1b... 150 1e-36
xla:397868 hdac1-a, HD1, HDM, MGC83956, ab21, gon-10, hdac1a, ... 149 2e-36
dre:192302 hdac1, MGC101582, chunp6919, hdac-1, mp:zf637-2-001... 149 4e-36
hsa:3065 HDAC1, DKFZp686H12203, GON-10, HD1, RPD3, RPD3L1; his... 147 7e-36
mmu:433759 Hdac1, HD1, Hdac1-ps, MGC102534, MGC118085, MommeD5... 146 2e-35
ath:AT4G38130 HD1; HD1 (HISTONE DEACETYLASE 1); basal transcri... 139 3e-33
sce:YNL330C RPD3, MOF6, REC3, SDI2, SDS6; Histone deacetylase;... 137 1e-32
ath:AT5G63110 HDA6; HDA6 (HISTONE DEACETYLASE 6); histone deac... 131 7e-31
cel:C53A5.3 hda-1; Histone DeAcetylase family member (hda-1); ... 130 1e-30
dre:393965 hdac3, MGC55927, zgc:55927; histone deacetylase 3 (... 129 3e-30
mmu:15183 Hdac3, AW537363; histone deacetylase 3 (EC:3.5.1.98)... 128 6e-30
hsa:8841 HDAC3, HD3, RPD3, RPD3-2; histone deacetylase 3 (EC:3... 128 6e-30
cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3); ... 125 3e-29
ath:AT3G44680 HDA9; HDA9 (HISTONE DEACETYLASE 9); histone deac... 121 6e-28
cpv:cgd6_1380 histone deacetylase ; K06067 histone deacetylase... 118 5e-27
ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deace... 113 2e-25
sce:YGL194C HOS2, RTL1; Histone deacetylase required for gene ... 96.3 2e-20
cel:C08B11.2 hda-2; Histone DeAcetylase family member (hda-2);... 90.1 2e-18
tgo:TGME49_049620 histone deacetylase, putative ; K06067 histo... 75.1 6e-14
dre:406740 hdac8, wu:fd19a02, zgc:66196; histone deacetylase 8... 58.9 5e-09
mmu:70315 Hdac8, 2610007D20Rik; histone deacetylase 8 (EC:3.5.... 54.7 8e-08
hsa:10013 HDAC6, FLJ16239, HD6; histone deacetylase 6 (EC:3.5.... 34.3 0.11
xla:444137 hdac8, MGC80565; histone deacetylase 8 (EC:3.5.1.98... 34.3 0.14
cel:C05D10.2 hypothetical protein; K08293 mitogen-activated pr... 31.2 1.0
sce:YPR068C HOS1; Hos1p; K11482 histone deacetylase HOS1 [EC:3... 30.8 1.3
xla:734402 hdac10, MGC115178; histone deacetylase 10 (EC:3.5.1... 30.8 1.5
mmu:18854 Pml, 1200009E24Rik, AI661194, Trim19; promyelocytic ... 30.4 2.0
cel:F09F3.13 srx-137; Serpentine Receptor, class X family memb... 29.6 2.7
pfa:PF10_0331 Sec1 family protein, putative 29.6 2.9
dre:327253 fd19e10, wu:fd19e10; zgc:55652 (EC:3.5.1.98); K1140... 29.3 3.9
ath:AT1G08460 HDA08; HDA08; histone deacetylase 28.9 5.2
mmu:574418 Serinc4, RP23-433P19.11; serine incorporator 4 28.1
> tgo:TGME49_027290 histone deacetylase, putative ; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=451
Score = 231 bits (589), Expect = 6e-61, Method: Compositional matrix adjust.
Identities = 107/118 (90%), Positives = 113/118 (95%), Gaps = 0/118 (0%)
Query 19 MALLGVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIE 78
MAL +RKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIE
Sbjct 1 MALSALRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIE 60
Query 79 PELCLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
PELCLFHS +YI+FLS+VSPENYKEF+LQLK+FNVGEATDCPVFDGLF FQQACAG S
Sbjct 61 PELCLFHSSDYISFLSSVSPENYKEFSLQLKNFNVGEATDCPVFDGLFTFQQACAGAS 118
> bbo:BBOV_III011020 17.m07949; histone deacetylase; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=449
Score = 201 bits (512), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 88/113 (77%), Positives = 102/113 (90%), Gaps = 0/113 (0%)
Query 24 VRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCL 83
+ KRV+YFYDPD+GSYYYGPGHPMKPQRIRMAHALVLSYDLY+HMEV+RPHK++EPEL
Sbjct 1 MEKRVSYFYDPDVGSYYYGPGHPMKPQRIRMAHALVLSYDLYRHMEVFRPHKAVEPELLA 60
Query 84 FHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
FH EY+ FLS VSP+NY++FA QLK FNVGEATDCPVFDGL+ FQQ+C+G S
Sbjct 61 FHDHEYLQFLSGVSPDNYRDFAYQLKRFNVGEATDCPVFDGLYVFQQSCSGAS 113
> tpv:TP02_0039 histone deacetylase; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=447
Score = 201 bits (510), Expect = 8e-52, Method: Compositional matrix adjust.
Identities = 86/111 (77%), Positives = 101/111 (90%), Gaps = 0/111 (0%)
Query 26 KRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFH 85
KRV+YFYDPD+GS+YYGPGHPMKPQRIRMAHALVLSYDLY+HME++RPHK++EPEL FH
Sbjct 3 KRVSYFYDPDVGSFYYGPGHPMKPQRIRMAHALVLSYDLYRHMEIFRPHKAVEPELLSFH 62
Query 86 SQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
EY++FLS VSPENY++F QLK FNVGEATDCPVFDGL+ FQQ+C+G S
Sbjct 63 DSEYVHFLSGVSPENYRDFTYQLKRFNVGEATDCPVFDGLYVFQQSCSGAS 113
> cpv:cgd6_80 RPD3/HD1 histone deacetylase ; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=460
Score = 195 bits (495), Expect = 4e-50, Method: Composition-based stats.
Identities = 85/115 (73%), Positives = 99/115 (86%), Gaps = 0/115 (0%)
Query 22 LGVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPEL 81
L + KRV+YFYD DIGSYYYGPGHPMKPQRIRMAH L+LSYDLYKHME+Y+PHKS + EL
Sbjct 15 LKMAKRVSYFYDGDIGSYYYGPGHPMKPQRIRMAHNLILSYDLYKHMEIYKPHKSPQSEL 74
Query 82 CLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
FH ++YINFLS+++P+N K+F LQLK FN+GE TDCPVFDGLF FQQ CAG S
Sbjct 75 VYFHEEDYINFLSSINPDNSKDFGLQLKRFNLGETTDCPVFDGLFEFQQICAGGS 129
> pfa:PFI1260c PfHDAC1; histone deacetylase; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=449
Score = 189 bits (480), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 86/115 (74%), Positives = 99/115 (86%), Gaps = 0/115 (0%)
Query 22 LGVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPEL 81
+ RK+VAYF+DPDIGSYYYG GHPMKPQRIRM H+L++SY+LYK+MEVYRPHKS EL
Sbjct 1 MSNRKKVAYFHDPDIGSYYYGAGHPMKPQRIRMTHSLIVSYNLYKYMEVYRPHKSDVNEL 60
Query 82 CLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
LFH EYI+FLS++S ENY+EF QLK FNVGEATDCPVFDGLF FQQ+CAG S
Sbjct 61 TLFHDYEYIDFLSSISLENYREFTYQLKRFNVGEATDCPVFDGLFQFQQSCAGAS 115
> xla:399252 hdac2, MGC81850; histone deacetylase 2 (EC:3.5.1.98);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 152 bits (385), Expect = 2e-37, Method: Composition-based stats.
Identities = 66/114 (57%), Positives = 84/114 (73%), Gaps = 2/114 (1%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G +K+V Y+YD DIG+YYYG GHPMKP RIRM H L+L+Y LY+ ME+YRPHK+ E+
Sbjct 7 GAKKKVCYYYDGDIGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKATAEEMT 66
Query 83 LFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
+HS EYI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q G S
Sbjct 67 KYHSDEYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSTGGS 118
> hsa:3066 HDAC2, HD2, RPD3, YAF1; histone deacetylase 2 (EC:3.5.1.98);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 151 bits (382), Expect = 5e-37, Method: Composition-based stats.
Identities = 66/114 (57%), Positives = 84/114 (73%), Gaps = 2/114 (1%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G +K+V Y+YD DIG+YYYG GHPMKP RIRM H L+L+Y LY+ ME+YRPHK+ E+
Sbjct 7 GGKKKVCYYYDGDIGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKATAEEMT 66
Query 83 LFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
+HS EYI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q G S
Sbjct 67 KYHSDEYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSTGGS 118
> mmu:15182 Hdac2, D10Wsu179e, YAF1, Yy1bp, mRPD3; histone deacetylase
2 (EC:3.5.1.98); K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 151 bits (382), Expect = 6e-37, Method: Composition-based stats.
Identities = 66/114 (57%), Positives = 84/114 (73%), Gaps = 2/114 (1%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G +K+V Y+YD DIG+YYYG GHPMKP RIRM H L+L+Y LY+ ME+YRPHK+ E+
Sbjct 7 GGKKKVCYYYDGDIGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKATAEEMT 66
Query 83 LFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
+HS EYI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q G S
Sbjct 67 KYHSDEYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSTGGS 118
> xla:379083 hdac1-b, HD1, MGC53583, RPD3, gon-10, hdac1, hdac1b,
rpd3l1; histone deacetylase 1 (EC:3.5.1.98); K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=480
Score = 150 bits (378), Expect = 1e-36, Method: Composition-based stats.
Identities = 64/114 (56%), Positives = 84/114 (73%), Gaps = 2/114 (1%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G +K+V Y+YD D+G+YYYG GHPMKP RIRM H L+L+Y LY+ ME+YRPHK+ E+
Sbjct 6 GTKKKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKASAEEMT 65
Query 83 LFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
+HS +YI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q G S
Sbjct 66 KYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSTGGS 117
> xla:397868 hdac1-a, HD1, HDM, MGC83956, ab21, gon-10, hdac1a,
rpd3, rpd3l1; histone deacetylase 1 (EC:3.5.1.98); K06067
histone deacetylase 1/2 [EC:3.5.1.98]
Length=480
Score = 149 bits (376), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 67/119 (56%), Positives = 89/119 (74%), Gaps = 3/119 (2%)
Query 19 MAL-LGVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSI 77
MAL LG +K+V Y+YD D+G+YYYG GHPMKP RIRM H L+L+Y LY+ ME++RPHK+
Sbjct 1 MALTLGTKKKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIFRPHKAS 60
Query 78 EPELCLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
++ +HS +YI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q AG S
Sbjct 61 AEDMTKYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSAGGS 117
> dre:192302 hdac1, MGC101582, chunp6919, hdac-1, mp:zf637-2-001987,
wu:fb19h11, wu:fi06f03, zgc:101582, zgc:65818; histone
deacetylase 1 (EC:3.5.1.98); K06067 histone deacetylase 1/2
[EC:3.5.1.98]
Length=480
Score = 149 bits (375), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 65/119 (54%), Positives = 87/119 (73%), Gaps = 2/119 (1%)
Query 18 AMALLGVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSI 77
A++ G +K+V Y+YD D+G+YYYG GHPMKP RIRM H L+L+Y LY+ ME+YRPHK+
Sbjct 2 ALSSQGTKKKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKAN 61
Query 78 EPELCLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
E+ +HS +YI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q G S
Sbjct 62 AEEMTKYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSTGGS 118
> hsa:3065 HDAC1, DKFZp686H12203, GON-10, HD1, RPD3, RPD3L1; histone
deacetylase 1 (EC:3.5.1.98); K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=482
Score = 147 bits (372), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 64/114 (56%), Positives = 84/114 (73%), Gaps = 2/114 (1%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G R++V Y+YD D+G+YYYG GHPMKP RIRM H L+L+Y LY+ ME+YRPHK+ E+
Sbjct 6 GTRRKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKANAEEMT 65
Query 83 LFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
+HS +YI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q G S
Sbjct 66 KYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSTGGS 117
> mmu:433759 Hdac1, HD1, Hdac1-ps, MGC102534, MGC118085, MommeD5,
RPD3; histone deacetylase 1 (EC:3.5.1.98); K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=482
Score = 146 bits (369), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 63/114 (55%), Positives = 84/114 (73%), Gaps = 2/114 (1%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G +++V Y+YD D+G+YYYG GHPMKP RIRM H L+L+Y LY+ ME+YRPHK+ E+
Sbjct 6 GTKRKVCYYYDGDVGNYYYGQGHPMKPHRIRMTHNLLLNYGLYRKMEIYRPHKANAEEMT 65
Query 83 LFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
+HS +YI FL ++ P+N E++ Q++ FNVGE DCPVFDGLF F Q G S
Sbjct 66 KYHSDDYIKFLRSIRPDNMSEYSKQMQRFNVGE--DCPVFDGLFEFCQLSTGGS 117
> ath:AT4G38130 HD1; HD1 (HISTONE DEACETYLASE 1); basal transcription
repressor/ histone deacetylase/ protein binding; K06067
histone deacetylase 1/2 [EC:3.5.1.98]
Length=469
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 62/114 (54%), Positives = 86/114 (75%), Gaps = 2/114 (1%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
GV+++V YFYDP++G+YYYG GHPMKP RIRM HAL+ Y L +HM+V +P + + +LC
Sbjct 14 GVKRKVCYFYDPEVGNYYYGQGHPMKPHRIRMTHALLAHYGLLQHMQVLKPFPARDRDLC 73
Query 83 LFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
FH+ +Y++FL +++PE ++ QLK FNVGE DCPVFDGL++F Q AG S
Sbjct 74 RFHADDYVSFLRSITPETQQDQIRQLKRFNVGE--DCPVFDGLYSFCQTYAGGS 125
> sce:YNL330C RPD3, MOF6, REC3, SDI2, SDS6; Histone deacetylase;
regulates transcription and silencing; plays a role in regulating
Ty1 transposition; K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=433
Score = 137 bits (345), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 61/112 (54%), Positives = 82/112 (73%), Gaps = 2/112 (1%)
Query 25 RKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLF 84
++RVAYFYD D+G+Y YG GHPMKP RIRMAH+L+++Y LYK ME+YR + + E+C F
Sbjct 18 KRRVAYFYDADVGNYAYGAGHPMKPHRIRMAHSLIMNYGLYKKMEIYRAKPATKQEMCQF 77
Query 85 HSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
H+ EYI+FLS V+P+N + F + FNVG+ DCPVFDGL+ + G S
Sbjct 78 HTDEYIDFLSRVTPDNLEMFKRESVKFNVGD--DCPVFDGLYEYCSISGGGS 127
> ath:AT5G63110 HDA6; HDA6 (HISTONE DEACETYLASE 6); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=471
Score = 131 bits (329), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 63/116 (54%), Positives = 85/116 (73%), Gaps = 4/116 (3%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G ++RV+YFY+P IG YYYG GHPMKP RIRMAH+L++ Y L++ +E+ RP + ++
Sbjct 16 GRKRRVSYFYEPTIGDYYYGQGHPMKPHRIRMAHSLIIHYHLHRRLEISRPSLADASDIG 75
Query 83 LFHSQEYINFLSAVSPENYKE--FALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
FHS EY++FL++VSPE+ + A L+ FNVGE DCPVFDGLF F +A AG S
Sbjct 76 RFHSPEYVDFLASVSPESMGDPSAARNLRRFNVGE--DCPVFDGLFDFCRASAGGS 129
> cel:C53A5.3 hda-1; Histone DeAcetylase family member (hda-1);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=461
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 59/112 (52%), Positives = 81/112 (72%), Gaps = 2/112 (1%)
Query 25 RKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLF 84
++RVAY+YD +IG+YYYG GH MKP RIRM H LVL+Y LY+++E++RP + ++ F
Sbjct 12 KRRVAYYYDSNIGNYYYGQGHVMKPHRIRMTHHLVLNYGLYRNLEIFRPFPASFEDMTRF 71
Query 85 HSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
HS EY+ FL + +P+N K F Q+ FNVGE DCP+FDGL+ F Q +G S
Sbjct 72 HSDEYMTFLKSANPDNLKSFNKQMLKFNVGE--DCPLFDGLYEFCQLSSGGS 121
> dre:393965 hdac3, MGC55927, zgc:55927; histone deacetylase 3
(EC:3.5.1.98); K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 129 bits (324), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 59/113 (52%), Positives = 79/113 (69%), Gaps = 2/113 (1%)
Query 24 VRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCL 83
+ R AYFYDPD+G+++YG GHPMKP R+ + H+LVL Y LYK M V++P+K+ + ++C
Sbjct 1 MTNRTAYFYDPDVGNFHYGAGHPMKPHRLSLTHSLVLHYGLYKKMMVFKPYKASQHDMCR 60
Query 84 FHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
FHS++YI+FL VSP N + F L +FNVG DCPVF GLF F G S
Sbjct 61 FHSEDYIDFLQKVSPNNMQGFTKSLNTFNVG--GDCPVFPGLFEFCSRYTGAS 111
> mmu:15183 Hdac3, AW537363; histone deacetylase 3 (EC:3.5.1.98);
K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 128 bits (321), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 59/111 (53%), Positives = 80/111 (72%), Gaps = 2/111 (1%)
Query 26 KRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFH 85
K VAYFYDPD+G+++YG GHPMKP R+ + H+LVL Y LYK M V++P+++ + ++C FH
Sbjct 3 KTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCRFH 62
Query 86 SQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
S++YI+FL VSP N + F L +FNVG+ DCPVF GLF F G S
Sbjct 63 SEDYIDFLQRVSPTNMQGFTKSLNAFNVGD--DCPVFPGLFEFCSRYTGAS 111
> hsa:8841 HDAC3, HD3, RPD3, RPD3-2; histone deacetylase 3 (EC:3.5.1.98);
K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 128 bits (321), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 59/111 (53%), Positives = 80/111 (72%), Gaps = 2/111 (1%)
Query 26 KRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFH 85
K VAYFYDPD+G+++YG GHPMKP R+ + H+LVL Y LYK M V++P+++ + ++C FH
Sbjct 3 KTVAYFYDPDVGNFHYGAGHPMKPHRLALTHSLVLHYGLYKKMIVFKPYQASQHDMCRFH 62
Query 86 SQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
S++YI+FL VSP N + F L +FNVG+ DCPVF GLF F G S
Sbjct 63 SEDYIDFLQRVSPTNMQGFTKSLNAFNVGD--DCPVFPGLFEFCSRYTGAS 111
> cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=465
Score = 125 bits (315), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 56/118 (47%), Positives = 81/118 (68%), Gaps = 3/118 (2%)
Query 19 MALLGVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIE 78
M+L + RV+Y+YD D G++YYG GHPMKP R+RM H+L+++Y LY+ + V RP ++
Sbjct 1 MSLQHSKSRVSYYYDGDFGNFYYGQGHPMKPHRVRMTHSLIVNYGLYRKLNVMRPARASF 60
Query 79 PELCLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAF-QQACAGT 135
E+ +HS +YINFL V +N F Q+ F+VGE DCPVFDG++ F Q +C G+
Sbjct 61 SEITRYHSDDYINFLRNVKSDNMSTFTDQMARFSVGE--DCPVFDGMYEFCQLSCGGS 116
> ath:AT3G44680 HDA9; HDA9 (HISTONE DEACETYLASE 9); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=426
Score = 121 bits (304), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 55/112 (49%), Positives = 77/112 (68%), Gaps = 2/112 (1%)
Query 25 RKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLF 84
+ +++YFYD D+GS Y+GP HPMKP R+ M H L+L+Y L+ MEVYRPHK+ E+ F
Sbjct 4 KDKISYFYDGDVGSVYFGPNHPMKPHRLCMTHHLILAYGLHSKMEVYRPHKAYPIEMAQF 63
Query 85 HSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
HS +Y+ FL ++PEN F ++ +N+GE DCPVF+ LF F Q AG +
Sbjct 64 HSPDYVEFLQRINPENQNLFPNEMARYNLGE--DCPVFEDLFEFCQLYAGGT 113
> cpv:cgd6_1380 histone deacetylase ; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=440
Score = 118 bits (296), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 54/115 (46%), Positives = 77/115 (66%), Gaps = 2/115 (1%)
Query 22 LGVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPEL 81
+G +K++AYFYD ++G+++YG GHPMKP R+RM H LV Y L + ++V P L
Sbjct 9 MGAKKKIAYFYDEEVGNFHYGLGHPMKPHRVRMTHDLVSQYGLLEKVDVMVPTPGTVESL 68
Query 82 CLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
FHS +Y++FL +V+ +N +++ L FNVGE DCPVFDGL+ F Q AG S
Sbjct 69 TRFHSNDYVDFLRSVNTDNMHDYSDHLARFNVGE--DCPVFDGLWEFCQLSAGGS 121
> ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=409
Score = 113 bits (283), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 57/118 (48%), Positives = 75/118 (63%), Gaps = 4/118 (3%)
Query 23 GVRKRVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
G ++RV+YFY+P IG YYYG P KPQRIR+ H L+LSY+L++HME+ P + +
Sbjct 7 GGKRRVSYFYEPMIGDYYYGVNQPTKPQRIRVTHNLILSYNLHRHMEINHPDLADASDFE 66
Query 83 LFHSQEYINFLSAVSPENYKE----FALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
FHS EYINFL +V+PE + + LK FNV D PVF LF + +A AG S
Sbjct 67 KFHSLEYINFLKSVTPETVTDPHPSVSENLKRFNVDVDWDGPVFHNLFDYCRAYAGGS 124
> sce:YGL194C HOS2, RTL1; Histone deacetylase required for gene
activation via specific deacetylation of lysines in H3 and
H4 histone tails; subunit of the Set3 complex, a meiotic-specific
repressor of sporulation specific genes that contains
deacetylase activity; K11483 histone deacetylase HOS2 [EC:3.5.1.98]
Length=452
Score = 96.3 bits (238), Expect = 2e-20, Method: Composition-based stats.
Identities = 46/111 (41%), Positives = 68/111 (61%), Gaps = 3/111 (2%)
Query 27 RVAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFHS 86
RV+Y ++ + Y+YG HPMKP R+ + LV SY L+K M++Y + EL FHS
Sbjct 27 RVSYHFNSKVSHYHYGVKHPMKPFRLMLTDHLVSSYGLHKIMDLYETRSATRDELLQFHS 86
Query 87 QEYINFLSAVSPENYKEFAL-QLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
++Y+NFLS VSPEN + L++FN+G+ DCP+F L+ + G S
Sbjct 87 EDYVNFLSKVSPENANKLPRGTLENFNIGD--DCPIFQNLYDYTTLYTGAS 135
> cel:C08B11.2 hda-2; Histone DeAcetylase family member (hda-2);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=507
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 49/119 (41%), Positives = 75/119 (63%), Gaps = 12/119 (10%)
Query 24 VRKR-VAYFYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELC 82
V+KR VAY+Y D+G ++YG HPMKPQR+ + + LV+SY++ K+M V K ++
Sbjct 26 VKKRNVAYYYHKDVGHFHYGQLHPMKPQRLVVCNDLVVSYEMPKYMTVVESPKLDAADIS 85
Query 83 LFHSQEYINFLSAVSPENYKEFALQ-----LKSFNVGEATDCPVFDGLFAFQQACAGTS 136
+FH+++Y+NFL V+P + L L+ FN+GE DCP+F GL+ + AG S
Sbjct 86 VFHTEDYVNFLQTVTP----KLGLTMPDDVLRQFNIGE--DCPIFAGLWDYCTLYAGGS 138
> tgo:TGME49_049620 histone deacetylase, putative ; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=734
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 53/81 (65%), Gaps = 1/81 (1%)
Query 31 FYDPDIGSYYYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFHSQEYI 90
F +IG+++YGP HPMKP R+RM H L+ +YDL + M++ +P K +L FH+ +Y+
Sbjct 120 FLSENIGNFHYGPSHPMKPHRVRMTHDLIQAYDLLQSMDIVKPLKPAVEDLTRFHANDYV 179
Query 91 NFLSAVSPENYKEFALQLKSF 111
+FL A S ++ E +L + F
Sbjct 180 DFLRA-SAQSRLEASLSPRPF 199
Score = 37.0 bits (84), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 18/33 (54%), Positives = 22/33 (66%), Gaps = 2/33 (6%)
Query 104 FALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
A Q+ FNVGE DCPVFDGL+ + Q +G S
Sbjct 385 LADQMARFNVGE--DCPVFDGLWEYCQTYSGGS 415
> dre:406740 hdac8, wu:fd19a02, zgc:66196; histone deacetylase
8 (EC:3.5.1.98); K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=378
Score = 58.9 bits (141), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 47/88 (53%), Gaps = 3/88 (3%)
Query 49 PQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFHSQEYINFLSAVSPENYKEFALQL 108
P R M H+L+ +Y L K+M V +PH + E+ +FH+ Y+ L +S + +
Sbjct 36 PNRASMVHSLIEAYGLLKYMRVVKPHVASIEEMAVFHTDSYLQHLHKISQDGDND---DP 92
Query 109 KSFNVGEATDCPVFDGLFAFQQACAGTS 136
+S + G DCPV +G+F + A G +
Sbjct 93 QSADFGLGYDCPVVEGIFDYAAAVGGAT 120
> mmu:70315 Hdac8, 2610007D20Rik; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=377
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 44/88 (50%), Gaps = 3/88 (3%)
Query 49 PQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFHSQEYINFLSAVSPENYKEFALQL 108
P+R M H+L+ +Y L+K M + +P + E+ FH+ Y+ L VS E ++
Sbjct 35 PKRASMVHSLIEAYALHKQMRIVKPKVASMEEMATFHTDAYLQHLQKVSQEGDEDHP--- 91
Query 109 KSFNVGEATDCPVFDGLFAFQQACAGTS 136
S G DCP +G+F + A G +
Sbjct 92 DSIEYGLGYDCPATEGIFDYAAAIGGGT 119
> hsa:10013 HDAC6, FLJ16239, HD6; histone deacetylase 6 (EC:3.5.1.98);
K11407 histone deacetylase 6/10 [EC:3.5.1.98]
Length=1215
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 31/115 (26%), Positives = 43/115 (37%), Gaps = 7/115 (6%)
Query 24 VRKRVAYFYDPDIGSY--YYGPGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPEL 81
++ R YD ++ ++ + HP PQRI + L P + E EL
Sbjct 477 LQSRTGLVYDQNMMNHCNLWDSHHPEVPQRILRIMCRLEELGLAGRCLTLTPRPATEAEL 536
Query 82 CLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQQACAGTS 136
HS EY+ L A E K L +S N CP FA Q G +
Sbjct 537 LTCHSAEYVGHLRAT--EKMKTRELHRESSNFDSIYICP---STFACAQLATGAA 586
> xla:444137 hdac8, MGC80565; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=325
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 31/67 (46%), Gaps = 3/67 (4%)
Query 70 VYRPHKSIEPELCLFHSQEYINFLSAVSPENYKEFALQLKSFNVGEATDCPVFDGLFAFQ 129
V +P + E+ FH+ Y+ L VS E + ++ G DCP+ +G++ +
Sbjct 4 VVKPKVASMEEMAAFHTDAYLQHLHKVSEEGDND---DPETLEYGLGYDCPITEGIYDYA 60
Query 130 QACAGTS 136
A G +
Sbjct 61 AAVGGAT 67
> cel:C05D10.2 hypothetical protein; K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=470
Score = 31.2 bits (69), Expect = 1.0, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 26/56 (46%), Gaps = 6/56 (10%)
Query 36 IGSYYYG------PGHPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFH 85
IGS+Y P P KP + + + + D+ + + ++ P K + E CL H
Sbjct 248 IGSHYAASVLEKMPQRPRKPLDLIITQSQTAAIDMVQRLLIFAPQKRLTVEQCLVH 303
> sce:YPR068C HOS1; Hos1p; K11482 histone deacetylase HOS1 [EC:3.5.1.98]
Length=470
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 29/45 (64%), Gaps = 1/45 (2%)
Query 50 QRIRMAHALVLSYDLYKHM-EVYRPHKSIEPELCLFHSQEYINFL 93
Q+ ++ ++L+ +YDL +H EV + + +L FHS+ YI++L
Sbjct 25 QKSQLTYSLINAYDLLQHFDEVLTFPYARKDDLLEFHSKSYIDYL 69
> xla:734402 hdac10, MGC115178; histone deacetylase 10 (EC:3.5.1.98);
K11407 histone deacetylase 6/10 [EC:3.5.1.98]
Length=683
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 20/80 (25%), Positives = 38/80 (47%), Gaps = 4/80 (5%)
Query 29 AYFYDPDIGSY---YYGPGHPMK-PQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLF 84
A YD ++ SY + P ++ P+R+ ++ + YDL K ++ + E+ L
Sbjct 6 ALVYDEEMMSYKLLWDDPECSIEVPERLSSSYKRLQDYDLVKRCIQLPVREATDEEITLV 65
Query 85 HSQEYINFLSAVSPENYKEF 104
HS +Y+ + + N KE
Sbjct 66 HSHDYLQVVKSTQTMNEKEL 85
> mmu:18854 Pml, 1200009E24Rik, AI661194, Trim19; promyelocytic
leukemia; K10054 probable transcription factor PML
Length=839
Score = 30.4 bits (67), Expect = 2.0, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 28/63 (44%), Gaps = 1/63 (1%)
Query 50 QRIRMAHALVLSYDLY-KHMEVYRPHKSIEPELCLFHSQEYINFLSAVSPENYKEFALQL 108
QRIR + ALV LY EV H + LC +E N + ++EF L L
Sbjct 328 QRIRTSGALVKRMKLYASDQEVLDMHSFLRKALCSLRQEEPQNQKVQLLTRGFEEFKLCL 387
Query 109 KSF 111
+ F
Sbjct 388 QDF 390
> cel:F09F3.13 srx-137; Serpentine Receptor, class X family member
(srx-137)
Length=309
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 15 LPAAMALLGVRKRVAYFYDPDIGSY 39
L LLG+RK AY YDP+I S+
Sbjct 136 LAITYTLLGLRKLCAYVYDPEIFSW 160
> pfa:PF10_0331 Sec1 family protein, putative
Length=663
Score = 29.6 bits (65), Expect = 2.9, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 75 KSIEPELCLFHSQEYINFLSAVSPENYKEFALQLKSFNV 113
K IE + + YINFLS VS EN++ FA + N+
Sbjct 99 KVIEDMVKNMYGSYYINFLSYVSQENFEYFASECVKNNI 137
> dre:327253 fd19e10, wu:fd19e10; zgc:55652 (EC:3.5.1.98); K11407
histone deacetylase 6/10 [EC:3.5.1.98]
Length=676
Score = 29.3 bits (64), Expect = 3.9, Method: Composition-based stats.
Identities = 14/64 (21%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 49 PQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFHSQEYINFLSAVSPENYKEFALQL 108
P+R+ +++ + ++ L + + ++ E E+ L HS+EY+ + N +E
Sbjct 30 PERLTVSYEALRTHGLAQRCKAVPVRQATEQEILLAHSEEYLEAVKQTPGMNVEELMAFS 89
Query 109 KSFN 112
K +N
Sbjct 90 KKYN 93
> ath:AT1G08460 HDA08; HDA08; histone deacetylase
Length=377
Score = 28.9 bits (63), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 45 HPMKPQRIRMAHALVLSYDLYKHMEVYRPHKSIEPELCLFHSQEYINFL 93
HP R+R +++ + H+ + +I EL +FH+ EYI L
Sbjct 39 HPENADRVRNMLSILRRGPIAPHVNWFTGLPAIVSELLMFHTSEYIEKL 87
> mmu:574418 Serinc4, RP23-433P19.11; serine incorporator 4
Length=492
Score = 28.1 bits (61), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 116 ATDCPVFDGLFAFQQACAGTSQF 138
++DCPV G A + CAGT+ F
Sbjct 102 SSDCPVLSGSGAVYRVCAGTATF 124
Lambda K H
0.326 0.141 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40