bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3413_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
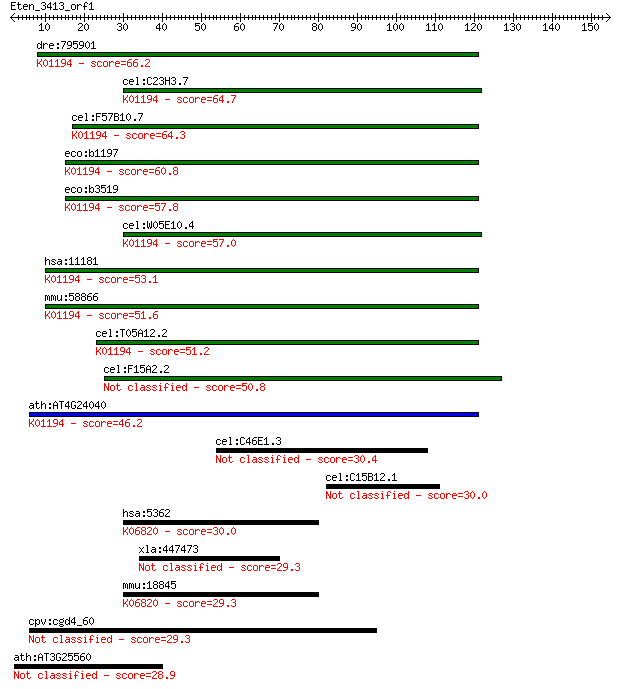
dre:795901 treh, si:ch211-147p17.2; trehalase (brush-border me... 66.2 4e-11
cel:C23H3.7 tre-5; TREhalase family member (tre-5); K01194 alp... 64.7 1e-10
cel:F57B10.7 tre-1; TREhalase family member (tre-1); K01194 al... 64.3 1e-10
eco:b1197 treA, ECK1185, JW1186, osmA; periplasmic trehalase (... 60.8 2e-09
eco:b3519 treF, ECK3504, JW3487; cytoplasmic trehalase (EC:3.2... 57.8 1e-08
cel:W05E10.4 tre-3; TREhalase family member (tre-3); K01194 al... 57.0 2e-08
hsa:11181 TREH, MGC129621, TRE, TREA; trehalase (brush-border ... 53.1 4e-07
mmu:58866 Treh, 2210412M19Rik; trehalase (brush-border membran... 51.6 1e-06
cel:T05A12.2 tre-2; TREhalase family member (tre-2); K01194 al... 51.2 1e-06
cel:F15A2.2 tre-4; TREhalase family member (tre-4) 50.8 2e-06
ath:AT4G24040 TRE1; TRE1 (TREHALASE 1); alpha,alpha-trehalase/... 46.2 4e-05
cel:C46E1.3 hypothetical protein 30.4 2.6
cel:C15B12.1 hypothetical protein 30.0 2.7
hsa:5362 PLXNA2, FLJ11751, FLJ30634, KIAA0463, OCT, PLXN2; ple... 30.0 3.0
xla:447473 ipo9, MGC81741; importin 9 29.3
mmu:18845 Plxna2, 2810428A13Rik, AA589422, AW457381, OCT, Plxn... 29.3 5.4
cpv:cgd4_60 extracellular membrane associated protein with a s... 29.3 5.7
ath:AT3G25560 NIK2; NIK2 (NSP-INTERACTING KINASE 2); ATP bindi... 28.9 6.5
> dre:795901 treh, si:ch211-147p17.2; trehalase (brush-border
membrane glycoprotein); K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=577
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/113 (34%), Positives = 61/113 (53%), Gaps = 0/113 (0%)
Query 8 SAQFLVEKSGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCM 67
+ Q+L + GL G+ + +GQQWD PN WPPL + +E + L +
Sbjct 433 AVQYLRDSGGLDYPNGIPTSLSDSGQQWDMPNAWPPLQHIIIEGLSGLHSAHAQELAFSL 492
Query 68 TDKYIFACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
++I +A+ + A+ EK + G G GG+Y+ Q+GFGW+ GVAL+LL
Sbjct 493 AQRWIQTNWRAFIKYEAMFEKYDVSGDGKPGGGGEYEVQLGFGWTNGVALQLL 545
> cel:C23H3.7 tre-5; TREhalase family member (tre-5); K01194 alpha,alpha-trehalase
[EC:3.2.1.28]
Length=674
Score = 64.7 bits (156), Expect = 1e-10, Method: Composition-based stats.
Identities = 32/93 (34%), Positives = 49/93 (52%), Gaps = 1/93 (1%)
Query 30 KTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACTQAYNRLGALPEKS 89
+T QQWD PN W P+ M +E + + L + +K++ Q +N + EK
Sbjct 514 ETNQQWDFPNGWSPMNHMIIEGLRKSNNPILQQKAFTLAEKWLETNMQTFNVSDEMWEKY 573
Query 90 NSLIS-GSCGSGGDYKCQIGFGWSIGVALELLF 121
N G +GG+Y+ Q GFGW+ G AL+L+F
Sbjct 574 NVKEPLGKLATGGEYEVQAGFGWTNGAALDLIF 606
> cel:F57B10.7 tre-1; TREhalase family member (tre-1); K01194
alpha,alpha-trehalase [EC:3.2.1.28]
Length=567
Score = 64.3 bits (155), Expect = 1e-10, Method: Composition-based stats.
Identities = 40/107 (37%), Positives = 56/107 (52%), Gaps = 8/107 (7%)
Query 17 GLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACT 76
GL S +S+TQ QWDK N WPP++ M +E D L+ + M ++
Sbjct 432 GLPTSLAMSSTQ-----QWDKENAWPPMIHMVIEGFRTTGDIKLMKVAEKMATSWLTGTY 486
Query 77 QAYNRLGALPEKSN---SLISGSCGSGGDYKCQIGFGWSIGVALELL 120
Q++ R A+ EK N S G GG+Y+ Q GFGW+ GV L+LL
Sbjct 487 QSFIRTHAMFEKYNVTPHTEETSGGGGGEYEVQTGFGWTNGVILDLL 533
> eco:b1197 treA, ECK1185, JW1186, osmA; periplasmic trehalase
(EC:3.2.1.28); K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=565
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 40/106 (37%), Positives = 57/106 (53%), Gaps = 3/106 (2%)
Query 15 KSGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFA 74
K+ L GL+ T VK+GQQWD PN W PL +A E + N + + I ++
Sbjct 427 KTHLLQPGGLNTTSVKSGQQWDAPNGWAPLQWVATEGLQNYGQKEVAMDISW---HFLTN 483
Query 75 CTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
Y+R L EK + +G+ G GG+Y Q GFGW+ GV L++L
Sbjct 484 VQHTYDREKKLVEKYDVSTTGTGGGGGEYPLQDGFGWTNGVTLKML 529
> eco:b3519 treF, ECK3504, JW3487; cytoplasmic trehalase (EC:3.2.1.28);
K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=549
Score = 57.8 bits (138), Expect = 1e-08, Method: Composition-based stats.
Identities = 36/113 (31%), Positives = 55/113 (48%), Gaps = 16/113 (14%)
Query 15 KSGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIIN-----LEDQALLSAIKCMTD 69
+S L G+ A++ +TG+QWDKPN W PL MA++ L D+ S +K +
Sbjct 440 RSRLLTPGGILASEYETGEQWDKPNGWAPLQWMAIQGFKMYGDDLLGDEIARSWLKTVNQ 499
Query 70 KYI--FACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
Y+ + Y+ +P + G GG+Y Q GFGW+ GV L+
Sbjct 500 FYLEQHKLIEKYHIADGVPRE---------GGGGEYPLQDGFGWTNGVVRRLI 543
> cel:W05E10.4 tre-3; TREhalase family member (tre-3); K01194
alpha,alpha-trehalase [EC:3.2.1.28]
Length=588
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 36/102 (35%), Positives = 51/102 (50%), Gaps = 19/102 (18%)
Query 30 KTGQQWDKPNCWPPLVQMAVEAI-----INLEDQALLSAIK-CMTDKYIFACT----QAY 79
++ QQWD PN W P M +E + ++D+ L A K M + +F T + Y
Sbjct 446 ESDQQWDFPNGWSPNNHMIIEGLRKSANPEMQDKGFLIASKWVMGNFRVFYETGHMWEKY 505
Query 80 NRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELLF 121
N +G+ P+ GSGG+Y Q GFGWS G L+LL
Sbjct 506 NVIGSYPQP---------GSGGEYDVQDGFGWSNGAILDLLL 538
> hsa:11181 TREH, MGC129621, TRE, TREA; trehalase (brush-border
membrane glycoprotein) (EC:3.2.1.28); K01194 alpha,alpha-trehalase
[EC:3.2.1.28]
Length=583
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 54/111 (48%), Gaps = 0/111 (0%)
Query 10 QFLVEKSGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTD 69
++L + L +G+ + KTGQQWD PN W PL + + + + +
Sbjct 437 KYLEDNRILTYQYGIPTSLQKTGQQWDFPNAWAPLQDLVIRGLAKAPLRRAQEVAFQLAQ 496
Query 70 KYIFACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
+I Y++ A+ EK + G G GG+Y+ Q GFGW+ GV L LL
Sbjct 497 NWIRTNFDVYSQKSAMYEKYDVSNGGQPGGGGEYEVQEGFGWTNGVVLMLL 547
> mmu:58866 Treh, 2210412M19Rik; trehalase (brush-border membrane
glycoprotein) (EC:3.2.1.28); K01194 alpha,alpha-trehalase
[EC:3.2.1.28]
Length=576
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 54/111 (48%), Gaps = 0/111 (0%)
Query 10 QFLVEKSGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTD 69
++L + L +G+ + TGQQWD PN W PL + + + +
Sbjct 434 KYLEDSKILTYQYGIPTSLRNTGQQWDFPNAWAPLQDLVIRGLAKSASPRTQEVAFQLAQ 493
Query 70 KYIFACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
+I + Y++ A+ EK + G G GG+Y+ Q GFGW+ G+AL LL
Sbjct 494 NWIKTNFKVYSQKSAMFEKYDISNGGHPGGGGEYEVQEGFGWTNGLALMLL 544
> cel:T05A12.2 tre-2; TREhalase family member (tre-2); K01194
alpha,alpha-trehalase [EC:3.2.1.28]
Length=585
Score = 51.2 bits (121), Expect = 1e-06, Method: Composition-based stats.
Identities = 34/103 (33%), Positives = 50/103 (48%), Gaps = 11/103 (10%)
Query 23 GLSATQVKTGQQWDKPNCWPPLVQMAVEAI--INLEDQALLSAIKCMTDKYIFACTQAYN 80
G+ + V +G+QWD PN WPP + +E + + E+ AL K + + T
Sbjct 422 GIPVSLVNSGEQWDFPNSWPPTTWVLLEGLRKVGQEELALSLVEKWVQKNFNMWRTSG-- 479
Query 81 RLGALPEKSNSLISGSC---GSGGDYKCQIGFGWSIGVALELL 120
G + EK N + C GG+Y Q GFGW+ GV L+ L
Sbjct 480 --GRMFEKYN--VVSPCFKVKGGGEYVMQEGFGWTNGVILDFL 518
> cel:F15A2.2 tre-4; TREhalase family member (tre-4)
Length=635
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 53/108 (49%), Gaps = 6/108 (5%)
Query 25 SATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACTQA-YNRLG 83
S+ ++ QQWD PN W P +++ + + L K ++I YN +
Sbjct 457 SSLPAQSTQQWDFPNVWAPNQHFVIQSFMACNNSFLQQEAKKQAMEFIETVYNGMYNPIA 516
Query 84 ALP----EKSNSL-ISGSCGSGGDYKCQIGFGWSIGVALELLFAKRGT 126
L EK ++ +G+ G+GG+Y Q GFGW+ G ++L++ R +
Sbjct 517 GLDGGVWEKYDARSTNGAPGAGGEYVVQEGFGWTNGAVMDLIWTLRDS 564
> ath:AT4G24040 TRE1; TRE1 (TREHALASE 1); alpha,alpha-trehalase/
trehalase (EC:3.2.1.28); K01194 alpha,alpha-trehalase [EC:3.2.1.28]
Length=626
Score = 46.2 bits (108), Expect = 4e-05, Method: Composition-based stats.
Identities = 40/122 (32%), Positives = 57/122 (46%), Gaps = 7/122 (5%)
Query 6 ISSAQFLVEK-------SGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQ 58
I+S + LV+K SGL G+ + +GQQWD PN W P +M V + +
Sbjct 492 INSDENLVKKVVTALKNSGLIAPAGILTSLTNSGQQWDSPNGWAPQQEMIVTGLGRSSVK 551
Query 59 ALLSAIKCMTDKYIFACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALE 118
+ + ++I + Y + G + EK G G GG+Y Q GFGWS GV L
Sbjct 552 EAKEMAEDIARRWIKSNYLVYKKSGTIHEKLKVTELGEYGGGGEYMPQTGFGWSNGVILA 611
Query 119 LL 120
L
Sbjct 612 FL 613
> cel:C46E1.3 hypothetical protein
Length=763
Score = 30.4 bits (67), Expect = 2.6, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 26/61 (42%), Gaps = 7/61 (11%)
Query 54 NLEDQALLSAIKCMTDKYIFACTQAYN-------RLGALPEKSNSLISGSCGSGGDYKCQ 106
NL+D A +S+IK D I+ YN GA P +N G CG G K
Sbjct 199 NLQDNASVSSIKTTDDYPIYNVLNDYNYSSGKTFHSGAFPIDANRKNFGFCGRSGSGKSS 258
Query 107 I 107
+
Sbjct 259 L 259
> cel:C15B12.1 hypothetical protein
Length=384
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 82 LGALPEKSNSLISGSCGSGGDYKCQIGFG 110
+G +P K+ +++ G CGSG +K G G
Sbjct 327 IGTIPTKNPNILVGGCGSGSGFKVAPGIG 355
> hsa:5362 PLXNA2, FLJ11751, FLJ30634, KIAA0463, OCT, PLXN2; plexin
A2; K06820 plexin A
Length=1894
Score = 30.0 bits (66), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 30 KTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACTQAY 79
KTG + D +C+PPL+ ++ L + I ++ + AC Y
Sbjct 84 KTGPEEDNKSCYPPLIVQPCSEVLTLTNNVNKLLIIDYSENRLLACGSLY 133
> xla:447473 ipo9, MGC81741; importin 9
Length=1033
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 11/36 (30%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 34 QWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTD 69
WD P WP L + +E +++ E A+ A++ +T+
Sbjct 132 HWDWPEAWPQLFNLLMEMLVSGEVNAVHGAMRVLTE 167
> mmu:18845 Plxna2, 2810428A13Rik, AA589422, AW457381, OCT, Plxn2,
mKIAA0463; plexin A2; K06820 plexin A
Length=1894
Score = 29.3 bits (64), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 22/50 (44%), Gaps = 0/50 (0%)
Query 30 KTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACTQAY 79
KTG + D C+PPL+ ++ L + I ++ + AC Y
Sbjct 84 KTGPEEDNKACYPPLIVQPCSEVLTLTNNVNKLLIIDYSENRLLACGSLY 133
> cpv:cgd4_60 extracellular membrane associated protein with a
signal peptide, an EGF domain followed by 10 transmembrane
domains
Length=789
Score = 29.3 bits (64), Expect = 5.7, Method: Composition-based stats.
Identities = 30/96 (31%), Positives = 39/96 (40%), Gaps = 13/96 (13%)
Query 6 ISSAQFLVEKSGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIK 65
IS A FL+ SGL D+ S T K G +D C P E + L L S +
Sbjct 18 ISQALFLIFLSGLIDAINSSDTPCKKGNYFDGNQCTP-----CPENTVGLTTN-LYSCVN 71
Query 66 CMTDKYIFACTQAY--NRLGALP-----EKSNSLIS 94
C + + T Y + LP E +NS IS
Sbjct 72 CPINSSTYGDTGLYSLDSCICLPGFFKLESTNSCIS 107
> ath:AT3G25560 NIK2; NIK2 (NSP-INTERACTING KINASE 2); ATP binding
/ protein binding / protein kinase/ protein serine/threonine
kinase
Length=635
Score = 28.9 bits (63), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 2 HKRQISSAQFLVEKSGLWDSWGLSATQVKTGQQWDKPN 39
H+ ++S ++E GL + W S+ + +T + + KPN
Sbjct 570 HRPKMSEVVRMLEGDGLVEKWEASSQRAETNRSYSKPN 607
Lambda K H
0.320 0.132 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40