bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3371_orf1
Length=343
Score E
Sequences producing significant alignments: (Bits) Value
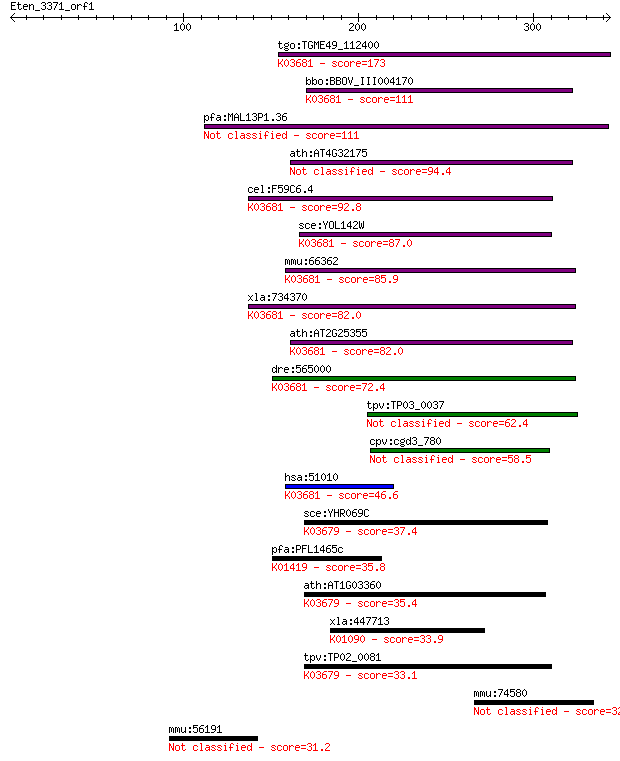
tgo:TGME49_112400 hypothetical protein ; K03681 exosome comple... 173 1e-42
bbo:BBOV_III004170 17.m07379; hypothetical protein; K03681 exo... 111 5e-24
pfa:MAL13P1.36 conserved Plasmodium protein, unknown function 111 5e-24
ath:AT4G32175 RNA binding 94.4 6e-19
cel:F59C6.4 exos-3; EXOSome (multiexonuclease complex) compone... 92.8 2e-18
sce:YOL142W RRP40; Exosome non-catalytic core component; invol... 87.0 1e-16
mmu:66362 Exosc3, 2310005D06Rik, AI593501, Rrp40; exosome comp... 85.9 2e-16
xla:734370 exosc3, MGC84934; exosome component 3; K03681 exoso... 82.0 3e-15
ath:AT2G25355 exonuclease-related; K03681 exosome complex comp... 82.0 3e-15
dre:565000 exosc3, MGC112345, im:7140537, zgc:112345; exosome ... 72.4 2e-12
tpv:TP03_0037 hypothetical protein 62.4 2e-09
cpv:cgd3_780 hypothetical protein 58.5 3e-08
hsa:51010 EXOSC3, MGC15120, MGC723, RP11-3J10.8, RRP40, Rrp40p... 46.6 1e-04
sce:YHR069C RRP4; Exosome non-catalytic core component; involv... 37.4 0.074
pfa:PFL1465c hslV, clpQ; Heat shock protein hslv (EC:3.4.25.-)... 35.8 0.24
ath:AT1G03360 ATRRP4; ATRRP4 (ARABIDOPSIS THALIANA RIBOSOMAL R... 35.4 0.31
xla:447713 ctdp1, MGC81710, ccfdn, fcp1; CTD (carboxy-terminal... 33.9 1.0
tpv:TP02_0081 hypothetical protein; K03679 exosome complex com... 33.1 1.8
mmu:74580 Pyroxd2, 3830409H07Rik, 4833409A17Rik; pyridine nucl... 32.7 2.1
mmu:56191 Tro, AA409408, Maged3, Maged3l, Tnn, Trol, magphinin... 31.2 6.7
> tgo:TGME49_112400 hypothetical protein ; K03681 exosome complex
component RRP40
Length=275
Score = 173 bits (438), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 90/190 (47%), Positives = 123/190 (64%), Gaps = 2/190 (1%)
Query 154 LPPHGPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATR 213
L P+ P+Y+ + RY P GD+VVGI+S K +E Y VNIR+ G L V+ FEGATR
Sbjct 84 LYPYSPSYI-RGSQKRYEPVEGDVVVGIVSGKKNEAYTVNIRARGDGSLPLVSSFEGATR 142
Query 214 RSKPQLSVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPF 273
R KP+L GS++L RV SP LGA++TC +C K WTS EKLLGEL GG +++V P
Sbjct 143 RHKPELRRGSLILCRVERASPELGAELTCIDPNCKKSWTSQEKLLGELEGGFVLDVPAPL 202
Query 274 ALTLLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCITAAAAGKTPAVL 333
A++L +L+ LG RL FEIA G NGRVW+RA+ ++ + N I A GK V+
Sbjct 203 AISLSSPHCFLLESLGERLAFEIASGANGRVWIRAKDAGDAILIGN-ILVACYGKERHVM 261
Query 334 QAIVDAVLQQ 343
QAI++ ++ +
Sbjct 262 QAIINTLVSK 271
> bbo:BBOV_III004170 17.m07379; hypothetical protein; K03681 exosome
complex component RRP40
Length=207
Score = 111 bits (277), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 56/157 (35%), Positives = 90/157 (57%), Gaps = 8/157 (5%)
Query 170 YCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGSVLLVRV 229
Y P++GD V+G++ +K S++Y ++I + A L V GF GAT+R++P ++ G V+ ++
Sbjct 33 YRPKIGDHVIGVVVAKNSDFYSLDIGGLTEAILPAVDGFRGATKRNRPNINEGDVIFCQI 92
Query 230 SHLSPL--LGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTL---LCHDSLV 284
+ P L +V+C K WT+ E G L GG + V IP++ L CH V
Sbjct 93 TKQYPRNELPVEVSCLDVDDMKQWTTKETYFGSLEGGFMFSVPIPYSYCLSGARCH---V 149
Query 285 LQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCI 321
L+ R +EIAVGLNGRVW+++ ++ +A I
Sbjct 150 LEKCAGRFKYEIAVGLNGRVWIKSTNEVDTILIARYI 186
> pfa:MAL13P1.36 conserved Plasmodium protein, unknown function
Length=245
Score = 111 bits (277), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 67/233 (28%), Positives = 117/233 (50%), Gaps = 7/233 (3%)
Query 112 HPSHPSGKDETQNHGPESAVEVEEAGPCVKARTASMMLPKHVLPPHGP-TYVCISTNNRY 170
H + K E N + E +++ + ++L P+ P Y ++T+ +Y
Sbjct 12 HMTIEKDKLEKMNENNFEKIYDNEYENIYRSKCSGILLK----TPYYPYKYDIMNTSYKY 67
Query 171 CPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGSVLLVRVS 230
P+VGD+V+GI+ SK +YY ++I + + F+ AT+ S P L G++L + V
Sbjct 68 IPKVGDLVIGIVKSKKLDYYQMDINCNCECIIHKIDSFKYATKSSFPNLLNGTLLYMVVE 127
Query 231 HLSPLLGADVT-CCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSLVLQLLG 289
++ + VT C +SS K W + E LGEL G + V I A +L+ +L L+G
Sbjct 128 KMNLENNSVVTSCINSSDVKSWINYENYLGELVDGFVFSVNIASAKSLIGDRCYILDLIG 187
Query 290 SRLPFEIAVGLNGRVWLRAETPTFSMRVANCITAAAAGKTPAVLQAIVDAVLQ 342
+ +EIAVG NGR+W++A P + + + + GKT A + + ++
Sbjct 188 QDIKYEIAVGHNGRIWIKANDP-LEINLIHSALKHSFGKTRAQMNVLWKSIYN 239
> ath:AT4G32175 RNA binding
Length=241
Score = 94.4 bits (233), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 60/163 (36%), Positives = 89/163 (54%), Gaps = 10/163 (6%)
Query 161 YVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPA-RLATVAGFEGATRRSKPQL 219
Y S++ RY PR D V+GI+ ++ + V+I+ GP L V FEG TRR+ P+
Sbjct 63 YWLESSHKRYIPRPDDHVLGIVVDSRADNFWVDIK--GPQLALLPVLAFEGGTRRNIPKF 120
Query 220 SVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLC 279
VG++L +RV + + +++C +S L G LR G + E + + LL
Sbjct 121 EVGTLLYLRVVKTNIGMNPELSCTEAS------GKAALFGPLRDGFMFETSTGLSRMLLN 174
Query 280 HDSL-VLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCI 321
+ V++ LG +L FEIAVGLNGRVW+ A P + VAN I
Sbjct 175 SPTRPVIEALGKKLSFEIAVGLNGRVWVNATAPRIVIIVANAI 217
> cel:F59C6.4 exos-3; EXOSome (multiexonuclease complex) component
family member (exos-3); K03681 exosome complex component
RRP40
Length=226
Score = 92.8 bits (229), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 52/176 (29%), Positives = 89/176 (50%), Gaps = 12/176 (6%)
Query 137 GPCVKARTASMMLPKHVLPPHGPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRS 196
G V+ +T P G ++ + + RY P+ GD V+ I++SK +++ ++I
Sbjct 24 GISVRGQTRIATQPGAFHNDDGKVWLNVHSK-RYIPQEGDRVIAIVTSKTGDFFRLDI-- 80
Query 197 VGPARLATV--AGFEGATRRSKPQLSVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSN 254
G A A + FEGAT+R++P L G ++ V +P A++TC
Sbjct 81 -GTAEYAMINFTNFEGATKRNRPNLKTGDIIYATVFDTTPRTEAELTCVDDE------KR 133
Query 255 EKLLGELRGGVIVEVAIPFALTLLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAET 310
+ +G+L GG + +V++ L+ +LQ +G FEI VG+NGR+W+ A T
Sbjct 134 ARGMGQLNGGFMFKVSLNHCRRLINPSCKILQTVGKFFKFEITVGMNGRIWISAST 189
> sce:YOL142W RRP40; Exosome non-catalytic core component; involved
in 3'-5' RNA processing and degradation in both the nucleus
and the cytoplasm; predicted to contain both S1 and KH
RNA binding domains; has similarity to human hRrp40p (EXOSC3)
(EC:3.1.13.-); K03681 exosome complex component RRP40
Length=240
Score = 87.0 bits (214), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 81/145 (55%), Gaps = 6/145 (4%)
Query 166 TNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGSVL 225
++ RY P V D V+G+I S+ Y V++++ + + F A+++++P L VG ++
Sbjct 60 SSKRYIPSVNDFVIGVIIGTFSDSYKVSLQNFSSSVSLSYMAFPNASKKNRPTLQVGDLV 119
Query 226 LVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSL-V 284
RV L A++ C S+ T + G L G+I++V FA LL ++ +
Sbjct 120 YARVCTAEKELEAEIECFDST-----TGRDAGFGILEDGMIIDVNFNFARQLLFNNDFPL 174
Query 285 LQLLGSRLPFEIAVGLNGRVWLRAE 309
L++L + FE+A+GLNG++W++ E
Sbjct 175 LKVLAAHTKFEVAIGLNGKIWVKCE 199
> mmu:66362 Exosc3, 2310005D06Rik, AI593501, Rrp40; exosome component
3; K03681 exosome complex component RRP40
Length=274
Score = 85.9 bits (211), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 53/170 (31%), Positives = 91/170 (53%), Gaps = 14/170 (8%)
Query 158 GPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKP 217
G Y S RY P GD V+GI+ +K + + V++ PA L+ +A FEGAT+R++P
Sbjct 96 GGVYWVDSQQKRYVPVKGDHVIGIVIAKSGDIFKVDVGGSEPASLSYLA-FEGATKRNRP 154
Query 218 QLSVGSVL----LVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPF 273
+ VG ++ +V + P + C SC + + ++G+ G++ +V +
Sbjct 155 NVQVGDLIYGQCVVANKDMEPEM-----VCIDSCGR--ANGMGVIGQ--DGLLFKVTLGL 205
Query 274 ALTLLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCITA 323
LL D ++Q LG P EI G+NGR+W++A+T ++ +AN + A
Sbjct 206 IRKLLAPDCEIVQELGKLYPLEIVFGMNGRIWVKAKTIQQTLILANVLEA 255
> xla:734370 exosc3, MGC84934; exosome component 3; K03681 exosome
complex component RRP40
Length=250
Score = 82.0 bits (201), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 51/189 (26%), Positives = 100/189 (52%), Gaps = 8/189 (4%)
Query 137 GPCVKARTASMMLPKHVLPPHGP--TYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNI 194
GP + + +++ K L H Y S RY P GD V+GI++++ + + V +
Sbjct 47 GPGLMRQEDDLLVNKCGLLRHRDPAVYWVDSQQKRYVPVKGDHVIGIVTTRAGDIFKVEV 106
Query 195 RSVGPARLATVAGFEGATRRSKPQLSVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSN 254
A L+ +A FEGAT++++P + G ++ + + + ++ C SS +
Sbjct 107 GGSEQASLSYLA-FEGATKKNRPNVKAGDLVYGQFIVANKDMEPEMACIDSSGK---ANG 162
Query 255 EKLLGELRGGVIVEVAIPFALTLLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAETPTFS 314
+ ++G+ G++ +V++ LL + ++Q L S P E+ VG+NGR+W++A+T +
Sbjct 163 KGVIGQ--DGLMFKVSLGLVRKLLTPQNDIIQELMSIFPLEVVVGMNGRIWVKAKTTQQT 220
Query 315 MRVANCITA 323
+ VAN + A
Sbjct 221 LIVANLLEA 229
> ath:AT2G25355 exonuclease-related; K03681 exosome complex component
RRP40
Length=213
Score = 82.0 bits (201), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 55/163 (33%), Positives = 81/163 (49%), Gaps = 10/163 (6%)
Query 161 YVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPA-RLATVAGFEGATRRSKPQL 219
Y S++ RY PR D V+GI+ E Y ++I+ GP L V FEGA RR+ P+
Sbjct 35 YWVESSHKRYVPRPEDHVLGIVVDCKGENYWIDIK--GPQLALLPVLAFEGANRRNYPKF 92
Query 220 SVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLC 279
V ++L RV + + +++C S G L+ G + E + + LL
Sbjct 93 EVSTLLYTRVVKTNTGMNPELSCVDESGKAAG------FGPLKDGFMFETSTGLSRMLLS 146
Query 280 HDSL-VLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCI 321
+ VL+ LG +L FE A GLNGR W+ A P + VAN +
Sbjct 147 SPTCPVLEALGKKLSFETAFGLNGRCWVHAAAPRIVIIVANAL 189
> dre:565000 exosc3, MGC112345, im:7140537, zgc:112345; exosome
component 3; K03681 exosome complex component RRP40
Length=247
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/174 (28%), Positives = 90/174 (51%), Gaps = 11/174 (6%)
Query 151 KHVLPPHGPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEG 210
KH P Y RY P G+ V+GI+++K + + V++ A L+ +A FEG
Sbjct 63 KHKQPN---MYWVNCQQRRYVPAKGESVIGIVTAKSGDVFKVDVGGSEQASLSYLA-FEG 118
Query 211 ATRRSKPQLSVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRG-GVIVEV 269
AT+R++P + VG ++ + + + + ++ C S C + +G G G++ +V
Sbjct 119 ATKRNRPNVQVGDLVFGQFTIANKDMEPELVCIDS-CGRA-----NGMGVFGGDGLLFKV 172
Query 270 AIPFALTLLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCITA 323
++ LL S ++ L P E+ VG+NGRVW++A T ++ ++N + A
Sbjct 173 SLGLVRRLLAPQSDLVSDLEKMFPCEMVVGMNGRVWVKARTVQHTLILSNLLEA 226
> tpv:TP03_0037 hypothetical protein
Length=143
Score = 62.4 bits (150), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/126 (23%), Positives = 63/126 (50%), Gaps = 7/126 (5%)
Query 205 VAGFEGATRRSKPQLSVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGG 264
+ GF G T++ KP L++G V+ V + L +++C + +K W++NE G L G
Sbjct 3 IDGFRGTTKKYKPNLTIGDVVFCYVYGMYDGL-IELSCVTMDDNKNWSTNETYFGPLSNG 61
Query 265 VIVEVAIPFALTLLCHDSLVLQLLGSRL------PFEIAVGLNGRVWLRAETPTFSMRVA 318
+ ++ + + + +++ +L L +EI +G NGRVW+ + ++++
Sbjct 62 FLTQLPLQHVQSYVNTVNIMFRLYNEELEVLRGLKYEIVLGFNGRVWIGNTSNELKLKIS 121
Query 319 NCITAA 324
I +
Sbjct 122 RFIKLS 127
> cpv:cgd3_780 hypothetical protein
Length=211
Score = 58.5 bits (140), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 57/133 (42%), Gaps = 31/133 (23%)
Query 207 GFEGATRRSKPQLSVGSVLLVRVSHLSPLLGA-DVTCCSSSCSKPWTSNEKLLGELRG-- 263
FEGAT+R+KP L G+ + R++++ G ++TC + K W++NE LG L+
Sbjct 31 AFEGATKRNKPNLVPGNYVCSRINYVDLESGEIELTCITPEEKKTWSNNENYLGVLKNSN 90
Query 264 ----------------------------GVIVEVAIPFALTLLCHDSLVLQLLGSRLPFE 295
G+ V V A LL S VL+ L +E
Sbjct 91 YKNTNIIEDYSKSDRSSLPGESQSCIKDGMTVTVPQSVAQILLADQSYVLEALSKYFAYE 150
Query 296 IAVGLNGRVWLRA 308
I VG NG +W A
Sbjct 151 ICVGQNGIIWFSA 163
> hsa:51010 EXOSC3, MGC15120, MGC723, RP11-3J10.8, RRP40, Rrp40p,
bA3J10.7, hRrp-40, hRrp40p, p10; exosome component 3; K03681
exosome complex component RRP40
Length=164
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 158 GPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKP 217
G Y S RY P GD V+GI+++K + + V++ PA L+ ++ FEGAT+R++P
Sbjct 97 GGVYWVDSQQKRYVPVKGDHVIGIVTAKSGDIFKVDVGGSEPASLSYLS-FEGATKRNRP 155
Query 218 QL 219
+
Sbjct 156 NV 157
> sce:YHR069C RRP4; Exosome non-catalytic core component; involved
in 3'-5' RNA processing and degradation in both the nucleus
and the cytoplasm; predicted to contain RNA binding domains;
has similarity to human hRrp4p (EXOSC2) (EC:3.1.13.-);
K03679 exosome complex component RRP4
Length=359
Score = 37.4 bits (85), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 61/145 (42%), Gaps = 19/145 (13%)
Query 169 RYCPRVGDIVVGIISSKGSEYYGVNIRSVGPA--RLATVAGFEGATRRS--KPQLSVGSV 224
RY P GD VVG I+ G++ + V+I A L +V G RR +L + S
Sbjct 103 RYAPETGDHVVGRIAEVGNKRWKVDIGGKQHAVLMLGSVNLPGGILRRKSESDELQMRSF 162
Query 225 LLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSLV 284
L LL A+V S + G+LR G+ +V P +L + +
Sbjct 163 L-----KEGDLLNAEVQSLFQDGSASLHTRSLKYGKLRNGMFCQV--PSSLIVRAKNHT- 214
Query 285 LQLLGSRLPFEIAV--GLNGRVWLR 307
LP I V G+NG +WLR
Sbjct 215 -----HNLPGNITVVLGVNGYIWLR 234
> pfa:PFL1465c hslV, clpQ; Heat shock protein hslv (EC:3.4.25.-);
K01419 ATP-dependent HslUV protease, peptidase subunit HslV
[EC:3.4.25.2]
Length=207
Score = 35.8 bits (81), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 5/62 (8%)
Query 151 KHVLPPHGPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEG 210
K ++P HG T +C+ NN C +GD +V S+G+ N + + + + GF G
Sbjct 30 KLIIPRHGTTILCVRKNNEVCL-IGDGMV----SQGTMIVKGNAKKIRRLKDNILMGFAG 84
Query 211 AT 212
AT
Sbjct 85 AT 86
> ath:AT1G03360 ATRRP4; ATRRP4 (ARABIDOPSIS THALIANA RIBOSOMAL
RNA PROCESSING 4); RNA binding / exonuclease; K03679 exosome
complex component RRP4
Length=322
Score = 35.4 bits (80), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 66/142 (46%), Gaps = 14/142 (9%)
Query 169 RYCPRVGDIVVGIISSKGSEYYGV--NIRSVGPARLATVAGFEGATRR--SKPQLSVGSV 224
RY P VGDIVVG + + + V N G L+++ +G RR S +L++ ++
Sbjct 88 RYKPEVGDIVVGRVIEVAQKRWRVELNFNQDGVLMLSSMNMPDGIQRRRTSVDELNMRNI 147
Query 225 LLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSLV 284
V H ++ A+V S + + G+L G +++V P+ + H
Sbjct 148 F---VEH--DVVCAEVRNFQHDGSLQLQARSQKYGKLEKGQLLKVD-PYLVKRSKHHFHY 201
Query 285 LQLLGSRLPFEIAVGLNGRVWL 306
++ LG ++ +G NG +W+
Sbjct 202 VESLG----IDLIIGCNGFIWV 219
> xla:447713 ctdp1, MGC81710, ccfdn, fcp1; CTD (carboxy-terminal
domain, RNA polymerase II, polypeptide A) phosphatase, subunit
1 (EC:3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=977
Score = 33.9 bits (76), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 25/88 (28%), Positives = 42/88 (47%), Gaps = 10/88 (11%)
Query 184 SKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGSVLLVRVSHLSPLLGADVTCC 243
S+ S+Y V + P RL + GA+ + +GSVL + V P+LGA+V
Sbjct 10 SQSSQYCEVRLSGPRPLRLVEWKVYAGAS------VQIGSVLALAV----PVLGAEVVRS 59
Query 244 SSSCSKPWTSNEKLLGELRGGVIVEVAI 271
+P + E+ + R GV+ E+ +
Sbjct 60 HEETGRPHRAPERKIKSDRAGVVQELCL 87
> tpv:TP02_0081 hypothetical protein; K03679 exosome complex component
RRP4
Length=263
Score = 33.1 bits (74), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 31/149 (20%), Positives = 60/149 (40%), Gaps = 23/149 (15%)
Query 169 RYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLA--TVAGFEGATRRSKPQLSVGSVLL 226
+Y ++GDIV+G++ S + +++ +V A+L+ V E A RR +
Sbjct 56 KYVGQIGDIVIGVVDSIQGNKWFLDVNAVELAQLSILQVNTEELANRRKIDE-------- 107
Query 227 VRVSHLSPLLGA-DVTCCSSSCSKP-----WTSNEKLLGELRGGVIVEVAIPFALTLLCH 280
+ +S L D+ C P + G+L G++V++ + H
Sbjct 108 -DIYEMSNLFNVNDIVSCEIQRISPTGTIMLQTRTSKYGKLENGILVKIKPNLVIRKGKH 166
Query 281 DSLVLQLLGSRLPFEIAVGLNGRVWLRAE 309
+ I +G NG +WL ++
Sbjct 167 ------IYDMECGVRIILGCNGYIWLTSQ 189
> mmu:74580 Pyroxd2, 3830409H07Rik, 4833409A17Rik; pyridine nucleotide-disulphide
oxidoreductase domain 2
Length=581
Score = 32.7 bits (73), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 38/73 (52%), Gaps = 11/73 (15%)
Query 266 IVEVAIPFAL--TLL---CHDSLVLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANC 320
++E+ IP +L TL CH V+ L P+ +A G+VW E T++ +V +C
Sbjct 433 MIELCIPSSLDPTLAPPGCH---VVSLFTQYTPYTLA---GGKVWNEQEKNTYADKVFDC 486
Query 321 ITAAAAGKTPAVL 333
I A A G +VL
Sbjct 487 IEAYAPGFKRSVL 499
> mmu:56191 Tro, AA409408, Maged3, Maged3l, Tnn, Trol, magphinin;
trophinin
Length=2087
Score = 31.2 bits (69), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query 92 ASASAAAATARSFQWSAVYVHPSHPSGKDETQNHGPESAVEVEEAGPCVK 141
AS+ A++R + SA V P P GK + N G SA E EA P ++
Sbjct 425 ASSRQTEASSRQIEASAAAVRPKKPRGK-KGNNKGSNSASEPSEAPPAIQ 473
Lambda K H
0.320 0.133 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14601168200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40