bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3350_orf4
Length=326
Score E
Sequences producing significant alignments: (Bits) Value
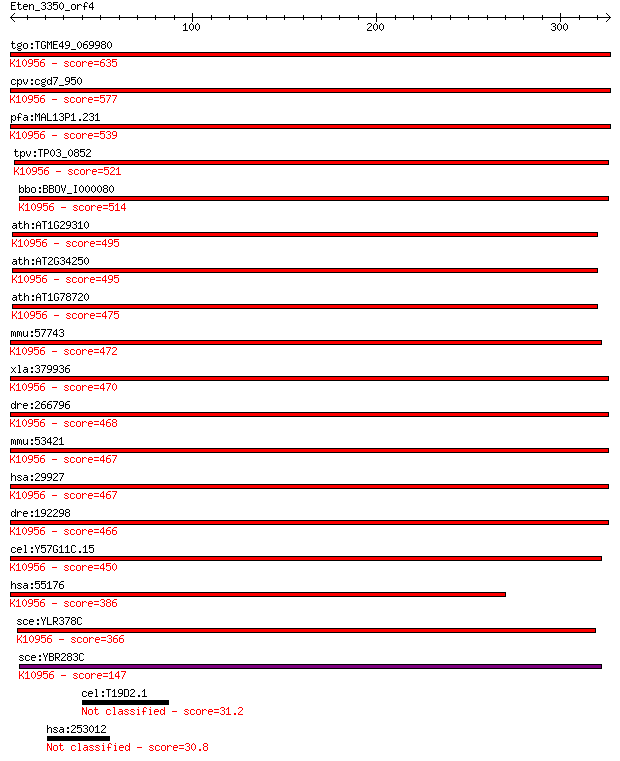
tgo:TGME49_069980 protein transport protein Sec61 alpha subuni... 635 0.0
cpv:cgd7_950 Sec61; signal peptide plus 9 transmembrane domain... 577 3e-164
pfa:MAL13P1.231 sec61, PF13_0244; Sec61 alpha subunit, PfSec61... 539 5e-153
tpv:TP03_0852 protein transport protein Sec61; K10956 protein ... 521 1e-147
bbo:BBOV_I000080 16.m00785; protein transport protein sec61; K... 514 2e-145
ath:AT1G29310 P-P-bond-hydrolysis-driven protein transmembrane... 495 9e-140
ath:AT2G34250 protein transport protein sec61, putative; K1095... 495 1e-139
ath:AT1G78720 protein transport protein sec61, putative; K1095... 475 1e-133
mmu:57743 Sec61a2; Sec61, alpha subunit 2 (S. cerevisiae); K10... 472 8e-133
xla:379936 sec61a1, MGC53497, sec61, sec61a, sec61alpha; Sec61... 470 3e-132
dre:266796 sec61al2, cb413, sec61b, wu:fb36f05, wu:fb36f12, wu... 468 1e-131
mmu:53421 Sec61a1, AA408394, AA410007, Sec61a; Sec61 alpha 1 s... 467 3e-131
hsa:29927 SEC61A1, HSEC61, SEC61, SEC61A; Sec61 alpha 1 subuni... 467 3e-131
dre:192298 sec61al1, chunp6898, fb62c11, sec61a, wu:fb62c11; S... 466 7e-131
cel:Y57G11C.15 hypothetical protein; K10956 protein transport ... 450 4e-126
hsa:55176 SEC61A2, FLJ10578; Sec61 alpha 2 subunit (S. cerevis... 386 8e-107
sce:YLR378C SEC61; Essential subunit of Sec61 complex (Sec61p,... 366 7e-101
sce:YBR283C SSH1; Ssh1p; K10956 protein transport protein SEC6... 147 4e-35
cel:T19D2.1 hypothetical protein 31.2 5.4
hsa:253012 HEPACAM2; HEPACAM family member 2 30.8
> tgo:TGME49_069980 protein transport protein Sec61 alpha subunit
isoform 1, putative ; K10956 protein transport protein SEC61
subunit alpha
Length=473
Score = 635 bits (1638), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 310/326 (95%), Positives = 321/326 (98%), Gaps = 0/326 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
A +A+LIILQLFF+GVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG
Sbjct 148 ATNAVLIILQLFFSGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 207
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
KGTEFEGALVALFHCLFTKS+N+ ALKEAFYRSNAPNITSLLAT+LVFLIVIYFQGFRVD
Sbjct 208 KGTEFEGALVALFHCLFTKSNNIVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVD 267
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFK NVLVNLLGQW
Sbjct 268 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKTNVLVNLLGQW 327
Query 181 QEVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSS 240
QEVD GGHSVPVGGIAYYISPPGSFG ILEDP+H+F+YITFVLVSCALFSKTWIEVSGSS
Sbjct 328 QEVDVGGHSVPVGGIAYYISPPGSFGDILEDPLHAFIYITFVLVSCALFSKTWIEVSGSS 387
Query 241 SRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG 300
+RDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG
Sbjct 388 ARDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG 447
Query 301 ILLAVTIIYQYYEMLAKEKEQGNSIF 326
ILLAVTIIYQYYEMLAKE+EQGNSIF
Sbjct 448 ILLAVTIIYQYYEMLAKEREQGNSIF 473
> cpv:cgd7_950 Sec61; signal peptide plus 9 transmembrane domain-containing
protein ; K10956 protein transport protein SEC61
subunit alpha
Length=473
Score = 577 bits (1486), Expect = 3e-164, Method: Compositional matrix adjust.
Identities = 274/326 (84%), Positives = 306/326 (93%), Gaps = 0/326 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
A +A+LII+QLFFAGVVVI+LDEL+QKGYGLGSGISLFIATNICETIVWKAFSPTTI TG
Sbjct 148 AWNAILIIIQLFFAGVVVILLDELMQKGYGLGSGISLFIATNICETIVWKAFSPTTINTG 207
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+GTEFEGA++ALFH LFTK ++AL+EAFYRS+A N+T+LLATVLVFLIVIYFQGFRVD
Sbjct 208 RGTEFEGAVIALFHLLFTKPDKISALREAFYRSHATNMTNLLATVLVFLIVIYFQGFRVD 267
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
LAVKYQ+VRGQQGS+PIKLFYTSNIPIILQTALVSNLYF SQLLYRRFK N+LVN+LGQW
Sbjct 268 LAVKYQKVRGQQGSFPIKLFYTSNIPIILQTALVSNLYFFSQLLYRRFKSNMLVNILGQW 327
Query 181 QEVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSS 240
QE+D GG S+PVGGIAYYISPP S ++ DP+H+F YI+FVL+SCALFSKTWIEVSGSS
Sbjct 328 QELDVGGQSIPVGGIAYYISPPNSLVDVVSDPIHTFFYISFVLISCALFSKTWIEVSGSS 387
Query 241 SRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG 300
++DVAKQLRDQQM+MKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG
Sbjct 388 AKDVAKQLRDQQMIMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG 447
Query 301 ILLAVTIIYQYYEMLAKEKEQGNSIF 326
ILLAVTII+QYYEM AKE+E GN +F
Sbjct 448 ILLAVTIIFQYYEMFAKERESGNIVF 473
> pfa:MAL13P1.231 sec61, PF13_0244; Sec61 alpha subunit, PfSec61;
K10956 protein transport protein SEC61 subunit alpha
Length=472
Score = 539 bits (1389), Expect = 5e-153, Method: Compositional matrix adjust.
Identities = 253/328 (77%), Positives = 300/328 (91%), Gaps = 2/328 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
G A++IILQLFFAGVVVI+LDELLQKGYGLGSGISLFIATNICETI+WK+FSPTTI T
Sbjct 145 TGHAIIIILQLFFAGVVVILLDELLQKGYGLGSGISLFIATNICETIMWKSFSPTTINTD 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
KG EFEGA+++L +CLFT+S+ ++ALK+AFYR++APN+T+LLAT+LVFLIVIY QGFRVD
Sbjct 205 KGIEFEGAIISLIYCLFTESNKISALKKAFYRTHAPNVTNLLATILVFLIVIYLQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L+VKYQ VRGQQG+YPIKLFYTSNIPIILQTALVSNLYF SQ+LY+RFK+++LVNLLGQW
Sbjct 265 LSVKYQSVRGQQGTYPIKLFYTSNIPIILQTALVSNLYFFSQILYKRFKNSILVNLLGQW 324
Query 181 QEVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSS 240
QEV++ G S+P+GG+AYYISPP SF I DP H+ VYI+FVLV+CA FSKTWIEVSGSS
Sbjct 325 QEVESSGTSIPIGGLAYYISPPNSFADITNDPFHTLVYISFVLVACAFFSKTWIEVSGSS 384
Query 241 SRDVAKQLRDQQMVMKGYRD--SSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
++DVAKQLRDQQ+ M+G+RD +SL +V NRYIPTAAAFGGMCIGALTI+ADFLGA+GSG
Sbjct 385 AKDVAKQLRDQQIGMRGFRDTPTSLTRVFNRYIPTAAAFGGMCIGALTILADFLGALGSG 444
Query 299 TGILLAVTIIYQYYEMLAKEKEQGNSIF 326
TGILLAVTIIYQ+YEML KE+E+ S+F
Sbjct 445 TGILLAVTIIYQFYEMLVKEQEKAASLF 472
> tpv:TP03_0852 protein transport protein Sec61; K10956 protein
transport protein SEC61 subunit alpha
Length=476
Score = 521 bits (1342), Expect = 1e-147, Method: Compositional matrix adjust.
Identities = 246/325 (75%), Positives = 293/325 (90%), Gaps = 3/325 (0%)
Query 3 SALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKG 62
++LIILQLFFAGVVVI+ DE+LQKGYGLGSGISLFIATNICETI+WKAFSPTTI T KG
Sbjct 150 KSVLIILQLFFAGVVVILFDEMLQKGYGLGSGISLFIATNICETILWKAFSPTTISTDKG 209
Query 63 TEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVDLA 122
TEFEGAL++LF+C FTK + L+A KEAFYR++APN+T+LLAT L+F+IVIY QGFRVDL+
Sbjct 210 TEFEGALISLFYCFFTKKNKLSAFKEAFYRNHAPNVTNLLATALIFVIVIYLQGFRVDLS 269
Query 123 VKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQWQE 182
VKYQ +RGQ+G+YPIKLFYTSNIPIILQTALVSNLYF SQL+YR+FK+N+ NLLGQWQE
Sbjct 270 VKYQSMRGQRGTYPIKLFYTSNIPIILQTALVSNLYFFSQLVYRKFKNNLFANLLGQWQE 329
Query 183 VDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSSSR 242
D G SVP+GG+AYY+SPP +F I+ DP+H+ +YITFVLVSCA+FSKTWIE+SGSS+R
Sbjct 330 TDH-GTSVPIGGLAYYLSPPSTFKDIVNDPLHTLLYITFVLVSCAVFSKTWIEISGSSAR 388
Query 243 DVAKQLRDQQMVMKGYRDS--SLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG 300
DVAKQLRDQ++ M G+RDS SL +V +RY+PTAAAFGGMCIGALTI+ADFLGA+GSGTG
Sbjct 389 DVAKQLRDQRIGMVGHRDSPPSLTKVFSRYVPTAAAFGGMCIGALTILADFLGALGSGTG 448
Query 301 ILLAVTIIYQYYEMLAKEKEQGNSI 325
ILLAVTIIYQYYE+L KE+E+ S+
Sbjct 449 ILLAVTIIYQYYEILVKEQERSGSL 473
> bbo:BBOV_I000080 16.m00785; protein transport protein sec61;
K10956 protein transport protein SEC61 subunit alpha
Length=480
Score = 514 bits (1324), Expect = 2e-145, Method: Compositional matrix adjust.
Identities = 243/322 (75%), Positives = 287/322 (89%), Gaps = 3/322 (0%)
Query 6 LIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEF 65
+IILQLF AGV+VI+ DE+LQKGYGLGSGISLFIATNICETI+WKAFSPTTI T KGTEF
Sbjct 157 VIILQLFMAGVIVILFDEMLQKGYGLGSGISLFIATNICETILWKAFSPTTISTDKGTEF 216
Query 66 EGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVDLAVKY 125
EGAL++LF+C FTK + L+A ++AFYRS+APN+T+LLAT L+F IVIY QGFRVDL +KY
Sbjct 217 EGALISLFYCFFTKGNKLSAFRDAFYRSHAPNVTNLLATALIFTIVIYLQGFRVDLPIKY 276
Query 126 QRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQWQEVDA 185
Q +RGQ+ +YPIKLFYTSNIPIILQTALVSNLYF SQL+YRRFK+N+ N+LGQWQE +
Sbjct 277 QNMRGQRSTYPIKLFYTSNIPIILQTALVSNLYFFSQLIYRRFKNNLFANILGQWQETEH 336
Query 186 GGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSSSRDVA 245
G SVPVGGIAYYISPP +F I+ DP+H+ VYITFVLVSCA+FSKTWIE+SGSS++DVA
Sbjct 337 GS-SVPVGGIAYYISPPINFKEIINDPIHTLVYITFVLVSCAVFSKTWIEISGSSAKDVA 395
Query 246 KQLRDQQMVMKGYRDS--SLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTGILL 303
+QLRDQ++ M GYRDS SL +V RYIPTAAAFGGMCIGALTI+ADFLGA+GSGTGILL
Sbjct 396 RQLRDQRIGMVGYRDSPPSLTKVFGRYIPTAAAFGGMCIGALTILADFLGALGSGTGILL 455
Query 304 AVTIIYQYYEMLAKEKEQGNSI 325
AVTIIYQY+EM+AKE+E+ S+
Sbjct 456 AVTIIYQYHEMMAKEQERSGSL 477
> ath:AT1G29310 P-P-bond-hydrolysis-driven protein transmembrane
transporter; K10956 protein transport protein SEC61 subunit
alpha
Length=475
Score = 495 bits (1274), Expect = 9e-140, Method: Compositional matrix adjust.
Identities = 234/318 (73%), Positives = 277/318 (87%), Gaps = 0/318 (0%)
Query 2 GSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGK 61
G+A+LIILQLFFAG++VI LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI TG+
Sbjct 149 GNAILIILQLFFAGIIVICLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINTGR 208
Query 62 GTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVDL 121
G EFEGA++ALFH L TKS+ +AAL++AFYR N PN+T+LLATVL+FLIVIYFQGFRV L
Sbjct 209 GAEFEGAVIALFHMLITKSNKVAALRQAFYRQNLPNVTNLLATVLIFLIVIYFQGFRVVL 268
Query 122 AVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQWQ 181
V+ + RGQQGSYPIKLFYTSN+PIILQ+ALVSNLYF+SQLLYR+F N VNLLGQW+
Sbjct 269 PVRSKSARGQQGSYPIKLFYTSNMPIILQSALVSNLYFISQLLYRKFSGNFFVNLLGQWK 328
Query 182 EVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSSS 241
E + G S+PV G+AY I+ P SF + P H+ YI F+L +CALFSKTWIEVSGSS+
Sbjct 329 ESEYSGQSIPVSGLAYLITAPASFADMAAHPFHALFYIVFMLTACALFSKTWIEVSGSSA 388
Query 242 RDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTGI 301
RDVAKQL++QQMVM G+R+S+L + LNRYIPTAAAFGG+CIGALT++ADF+GAIGSGTGI
Sbjct 389 RDVAKQLKEQQMVMPGHRESNLQKELNRYIPTAAAFGGVCIGALTVLADFMGAIGSGTGI 448
Query 302 LLAVTIIYQYYEMLAKEK 319
LLAVTIIYQY+E KEK
Sbjct 449 LLAVTIIYQYFETFEKEK 466
> ath:AT2G34250 protein transport protein sec61, putative; K10956
protein transport protein SEC61 subunit alpha
Length=475
Score = 495 bits (1274), Expect = 1e-139, Method: Compositional matrix adjust.
Identities = 234/318 (73%), Positives = 277/318 (87%), Gaps = 0/318 (0%)
Query 2 GSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGK 61
G+A+LIILQLFFAG++VI LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI TG+
Sbjct 149 GNAILIILQLFFAGIIVICLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINTGR 208
Query 62 GTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVDL 121
G EFEGA++ALFH L TKS+ +AAL++AFYR N PN+T+LLATVL+FLIVIYFQGFRV L
Sbjct 209 GAEFEGAVIALFHMLITKSNKVAALRQAFYRQNLPNVTNLLATVLIFLIVIYFQGFRVVL 268
Query 122 AVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQWQ 181
V+ + RGQQGSYPIKLFYTSN+PIILQ+ALVSNLYF+SQLLYR+F N VNLLGQW+
Sbjct 269 PVRSKNARGQQGSYPIKLFYTSNMPIILQSALVSNLYFISQLLYRKFSGNFFVNLLGQWK 328
Query 182 EVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSSS 241
E + G S+PV G+AY I+ P SF + P H+ YI F+L +CALFSKTWIEVSGSS+
Sbjct 329 ESEYSGQSIPVSGLAYLITAPASFSDMAAHPFHALFYIVFMLTACALFSKTWIEVSGSSA 388
Query 242 RDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTGI 301
RDVAKQL++QQMVM G+R+S+L + LNRYIPTAAAFGG+CIGALT++ADF+GAIGSGTGI
Sbjct 389 RDVAKQLKEQQMVMPGHRESNLQKELNRYIPTAAAFGGVCIGALTVLADFMGAIGSGTGI 448
Query 302 LLAVTIIYQYYEMLAKEK 319
LLAVTIIYQY+E KEK
Sbjct 449 LLAVTIIYQYFETFEKEK 466
> ath:AT1G78720 protein transport protein sec61, putative; K10956
protein transport protein SEC61 subunit alpha
Length=475
Score = 475 bits (1222), Expect = 1e-133, Method: Compositional matrix adjust.
Identities = 220/318 (69%), Positives = 272/318 (85%), Gaps = 0/318 (0%)
Query 2 GSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGK 61
G+A+LII+QL FA ++V+ LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI +G+
Sbjct 149 GNAILIIVQLCFAAIIVLCLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINSGR 208
Query 62 GTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVDL 121
G +FEGA++ALFH L T++ + AL+EAF+R N PN+T+L ATVL+FLIVIYFQGFRV L
Sbjct 209 GAQFEGAVIALFHLLITRTDKVRALREAFFRQNLPNVTNLHATVLIFLIVIYFQGFRVVL 268
Query 122 AVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQWQ 181
V+ + RGQ+GSYPIKLFYTSN+PIILQ+ALVSN+YF+SQ+LYR+F N LVNL+G W+
Sbjct 269 PVRSKNARGQRGSYPIKLFYTSNMPIILQSALVSNIYFISQILYRKFGGNFLVNLIGTWK 328
Query 182 EVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSSS 241
E + G S+PVGGIAYYI+ P S + P H+ Y+ F+L +CALFSKTWIEVSGSS+
Sbjct 329 ESEYSGQSIPVGGIAYYITAPSSLAEMATHPFHALFYLVFMLAACALFSKTWIEVSGSSA 388
Query 242 RDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTGI 301
+DVA+QLR+QQMVM G+RDS+L + LNRYIPTAAAFGG+CIGALT++AD +GAIGSGTGI
Sbjct 389 KDVARQLREQQMVMPGHRDSNLQKELNRYIPTAAAFGGLCIGALTVLADLMGAIGSGTGI 448
Query 302 LLAVTIIYQYYEMLAKEK 319
LLAVTIIYQY+E KEK
Sbjct 449 LLAVTIIYQYFETFEKEK 466
> mmu:57743 Sec61a2; Sec61, alpha subunit 2 (S. cerevisiae); K10956
protein transport protein SEC61 subunit alpha
Length=476
Score = 472 bits (1215), Expect = 8e-133, Method: Compositional matrix adjust.
Identities = 235/323 (72%), Positives = 276/323 (85%), Gaps = 2/323 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLII+QLF AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTI TG
Sbjct 145 AGICLLIIIQLFVAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTINTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+GTEFEGA++ALFH L T++ + AL+EAFYR N PN+ +L+ATV VF +VIYFQGFRVD
Sbjct 205 RGTEFEGAVIALFHLLATRTDKVRALREAFYRQNLPNLMNLIATVFVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ SYPIKLFYTSNIPIILQ+ALVSNLY +SQ+L RF N LVNLLGQW
Sbjct 265 LPIKSARYRGQYSSYPIKLFYTSNIPIILQSALVSNLYVISQMLSVRFSGNFLVNLLGQW 324
Query 181 QEVDAGG--HSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
+V GG S PVGG+ YY+SPP S G+I EDPVH VYI F+L SCA FSKTWIEVSG
Sbjct 325 ADVSGGGPARSYPVGGLCYYLSPPESMGAIFEDPVHVVVYIIFMLGSCAFFSKTWIEVSG 384
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
SS++DVAKQL++QQMVM+G+RD+S+V LNRYIPTAAAFGG+CIGAL+++ADFLGAIGSG
Sbjct 385 SSAKDVAKQLKEQQMVMRGHRDTSMVHELNRYIPTAAAFGGLCIGALSVLADFLGAIGSG 444
Query 299 TGILLAVTIIYQYYEMLAKEKEQ 321
TGILLAVTIIYQY+E+ KE+ +
Sbjct 445 TGILLAVTIIYQYFEIFVKEQAE 467
> xla:379936 sec61a1, MGC53497, sec61, sec61a, sec61alpha; Sec61
alpha 1 subunit; K10956 protein transport protein SEC61 subunit
alpha
Length=476
Score = 470 bits (1210), Expect = 3e-132, Method: Compositional matrix adjust.
Identities = 229/327 (70%), Positives = 281/327 (85%), Gaps = 2/327 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLI +QLF AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG
Sbjct 145 AGICLLITIQLFVAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTVNTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+GTEFEGA++ALFH L T+S + AL+EAFYR N PN+T+L+AT+ VF +VIYFQGFRVD
Sbjct 205 RGTEFEGAIIALFHLLATRSDKVRALREAFYRQNLPNLTNLIATIFVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ +YPIKLFYTSNIPIILQ+ALVSNLY +SQ+L RF N+LV+LLG W
Sbjct 265 LPIKSARYRGQYNTYPIKLFYTSNIPIILQSALVSNLYVISQMLSARFSGNLLVSLLGTW 324
Query 181 QEVDAGG--HSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
+ +GG + PVGG+ YY+SPP SFGS+L+DPVH+ +YI F+L SCA FSKTWIEVSG
Sbjct 325 SDTSSGGPARAYPVGGLCYYLSPPESFGSVLDDPVHAVIYIVFMLGSCAFFSKTWIEVSG 384
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
SS++DVAKQL++QQMVM+G+R++S+V LNRYIPTAAAFGG+CIGAL+++ADFLGAIGSG
Sbjct 385 SSAKDVAKQLKEQQMVMRGHRETSMVHELNRYIPTAAAFGGLCIGALSVLADFLGAIGSG 444
Query 299 TGILLAVTIIYQYYEMLAKEKEQGNSI 325
TGILLAVTIIYQY+E+ KE+ + S+
Sbjct 445 TGILLAVTIIYQYFEIFVKEQSEMGSM 471
> dre:266796 sec61al2, cb413, sec61b, wu:fb36f05, wu:fb36f12,
wu:fb37d04, wu:fi26f11; Sec61 alpha like 2; K10956 protein transport
protein SEC61 subunit alpha
Length=476
Score = 468 bits (1204), Expect = 1e-131, Method: Compositional matrix adjust.
Identities = 227/327 (69%), Positives = 278/327 (85%), Gaps = 2/327 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLII+QLF AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG
Sbjct 145 AGICLLIIIQLFVAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTVNTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+GTEFEGA++ALFH L T++ + AL+EAFYR N PN+ +L+AT+ VF +VIYFQGFRVD
Sbjct 205 RGTEFEGAIIALFHLLATRTDKVRALREAFYRQNLPNLMNLIATIFVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ +YPIKLFYTSNIPIILQ+ALVSNLY +SQ+L RF N LVNLLG W
Sbjct 265 LPIKSARYRGQYNTYPIKLFYTSNIPIILQSALVSNLYVISQMLSTRFSGNFLVNLLGTW 324
Query 181 QEVDAGG--HSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
+ GG + PVGG+ YY+SPP SFG++LEDP+H+ +YI F+L SCA FSKTWIEVSG
Sbjct 325 SDTSTGGPARAYPVGGLCYYLSPPESFGTVLEDPIHAIIYIIFMLGSCAFFSKTWIEVSG 384
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
SS++DVAKQL++QQMVM+G+R++S+V LNRYIPTAAAFGG+CIG L+++ADFLGAIGSG
Sbjct 385 SSAKDVAKQLKEQQMVMRGHRETSMVHELNRYIPTAAAFGGLCIGGLSVMADFLGAIGSG 444
Query 299 TGILLAVTIIYQYYEMLAKEKEQGNSI 325
TGILLAVTIIYQY+E+ KE+ + S+
Sbjct 445 TGILLAVTIIYQYFEIFVKEQSEVGSV 471
> mmu:53421 Sec61a1, AA408394, AA410007, Sec61a; Sec61 alpha 1
subunit (S. cerevisiae); K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 467 bits (1201), Expect = 3e-131, Method: Compositional matrix adjust.
Identities = 228/327 (69%), Positives = 279/327 (85%), Gaps = 2/327 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLI +QLF AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG
Sbjct 145 AGICLLITIQLFVAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTVNTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+G EFEGA++ALFH L T++ + AL+EAFYR N PN+ +L+AT+ VF +VIYFQGFRVD
Sbjct 205 RGMEFEGAIIALFHLLATRTDKVRALREAFYRQNLPNLMNLIATIFVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ +YPIKLFYTSNIPIILQ+ALVSNLY +SQ+L RF N+LV+LLG W
Sbjct 265 LPIKSARYRGQYNTYPIKLFYTSNIPIILQSALVSNLYVISQMLSARFSGNLLVSLLGTW 324
Query 181 QEVDAGG--HSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
+ +GG + PVGG+ YY+SPP SFGS+LEDPVH+ VYI F+L SCA FSKTWIEVSG
Sbjct 325 SDTSSGGPARAYPVGGLCYYLSPPESFGSVLEDPVHAVVYIVFMLGSCAFFSKTWIEVSG 384
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
SS++DVAKQL++QQMVM+G+R++S+V LNRYIPTAAAFGG+CIGAL+++ADFLGAIGSG
Sbjct 385 SSAKDVAKQLKEQQMVMRGHRETSMVHELNRYIPTAAAFGGLCIGALSVLADFLGAIGSG 444
Query 299 TGILLAVTIIYQYYEMLAKEKEQGNSI 325
TGILLAVTIIYQY+E+ KE+ + S+
Sbjct 445 TGILLAVTIIYQYFEIFVKEQSEVGSM 471
> hsa:29927 SEC61A1, HSEC61, SEC61, SEC61A; Sec61 alpha 1 subunit
(S. cerevisiae); K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 467 bits (1201), Expect = 3e-131, Method: Compositional matrix adjust.
Identities = 228/327 (69%), Positives = 279/327 (85%), Gaps = 2/327 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLI +QLF AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG
Sbjct 145 AGICLLITIQLFVAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTVNTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+G EFEGA++ALFH L T++ + AL+EAFYR N PN+ +L+AT+ VF +VIYFQGFRVD
Sbjct 205 RGMEFEGAIIALFHLLATRTDKVRALREAFYRQNLPNLMNLIATIFVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ +YPIKLFYTSNIPIILQ+ALVSNLY +SQ+L RF N+LV+LLG W
Sbjct 265 LPIKSARYRGQYNTYPIKLFYTSNIPIILQSALVSNLYVISQMLSARFSGNLLVSLLGTW 324
Query 181 QEVDAGG--HSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
+ +GG + PVGG+ YY+SPP SFGS+LEDPVH+ VYI F+L SCA FSKTWIEVSG
Sbjct 325 SDTSSGGPARAYPVGGLCYYLSPPESFGSVLEDPVHAVVYIVFMLGSCAFFSKTWIEVSG 384
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
SS++DVAKQL++QQMVM+G+R++S+V LNRYIPTAAAFGG+CIGAL+++ADFLGAIGSG
Sbjct 385 SSAKDVAKQLKEQQMVMRGHRETSMVHELNRYIPTAAAFGGLCIGALSVLADFLGAIGSG 444
Query 299 TGILLAVTIIYQYYEMLAKEKEQGNSI 325
TGILLAVTIIYQY+E+ KE+ + S+
Sbjct 445 TGILLAVTIIYQYFEIFVKEQSEVGSM 471
> dre:192298 sec61al1, chunp6898, fb62c11, sec61a, wu:fb62c11;
Sec61 alpha like 1; K10956 protein transport protein SEC61
subunit alpha
Length=476
Score = 466 bits (1198), Expect = 7e-131, Method: Compositional matrix adjust.
Identities = 228/327 (69%), Positives = 278/327 (85%), Gaps = 2/327 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLII+QLF AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG
Sbjct 145 AGICLLIIIQLFVAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTVNTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+GTEFEGA++ALFH L T++ + AL+EAFYR N PN+ +L+ATV VF +VIYFQGFRVD
Sbjct 205 RGTEFEGAIIALFHLLATRTDKVRALREAFYRQNLPNLMNLIATVFVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ +YPIKLFYTSNIPIILQ+ALVSNLY +SQ+L R N LVNLLG W
Sbjct 265 LPIKSARYRGQYNTYPIKLFYTSNIPIILQSALVSNLYVISQMLSTRSSGNFLVNLLGTW 324
Query 181 QEVDAGG--HSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
+ +GG + PVGG+ YY+SPP SFGS+L+DPVH+ +YI F+L SCA FSKTWIEVSG
Sbjct 325 SDTSSGGPARAYPVGGLCYYLSPPESFGSVLDDPVHAVIYIVFMLGSCAFFSKTWIEVSG 384
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
SS++DVAKQL++QQMVM+G+R++S+V LNRYIPTAAAFGG+CIG L+++ADFLGAIGSG
Sbjct 385 SSAKDVAKQLKEQQMVMRGHRETSMVHELNRYIPTAAAFGGLCIGGLSVMADFLGAIGSG 444
Query 299 TGILLAVTIIYQYYEMLAKEKEQGNSI 325
TGILLAVTIIYQY+E+ KE+ + S+
Sbjct 445 TGILLAVTIIYQYFEIFVKEQSEVGSM 471
> cel:Y57G11C.15 hypothetical protein; K10956 protein transport
protein SEC61 subunit alpha
Length=473
Score = 450 bits (1157), Expect = 4e-126, Method: Compositional matrix adjust.
Identities = 219/321 (68%), Positives = 268/321 (83%), Gaps = 1/321 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLI++QL AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSP T+ TG
Sbjct 145 AGICLLIVVQLVIAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPATMNTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+GTEFEGA++ALFH L T+S + AL+EAFYR N PN+ +L+AT LVF +VIYFQGFRVD
Sbjct 205 RGTEFEGAVIALFHLLATRSDKVRALREAFYRQNLPNLMNLMATFLVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ SYPIKLFYTSNIPIILQ+ALVSNLY +SQ+L +F N +NLLG W
Sbjct 265 LPIKSARYRGQYSSYPIKLFYTSNIPIILQSALVSNLYVISQMLAGKFGGNFFINLLGTW 324
Query 181 QEVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSS 240
+ + G S P GG+ YY+SPP S G I EDP+H +YI F+L SCA FSKTWI+VSGSS
Sbjct 325 SD-NTGYRSYPTGGLCYYLSPPESLGHIFEDPIHCIIYIVFMLGSCAFFSKTWIDVSGSS 383
Query 241 SRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTG 300
++DVAKQL++QQMVM+G+R+ S++ LNRYIPTAAAFGG+CIGAL++ ADF+GAIGSGTG
Sbjct 384 AKDVAKQLKEQQMVMRGHREKSMIHELNRYIPTAAAFGGLCIGALSVTADFMGAIGSGTG 443
Query 301 ILLAVTIIYQYYEMLAKEKEQ 321
ILLAVTIIYQY+E+ KE+++
Sbjct 444 ILLAVTIIYQYFEIFVKEQQE 464
> hsa:55176 SEC61A2, FLJ10578; Sec61 alpha 2 subunit (S. cerevisiae);
K10956 protein transport protein SEC61 subunit alpha
Length=437
Score = 386 bits (991), Expect = 8e-107, Method: Compositional matrix adjust.
Identities = 194/271 (71%), Positives = 227/271 (83%), Gaps = 2/271 (0%)
Query 1 AGSALLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTG 60
AG LLII+QLF AG++V++LDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTI TG
Sbjct 145 AGICLLIIIQLFVAGLIVLLLDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTINTG 204
Query 61 KGTEFEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVD 120
+GTEFEGA++ALFH L T++ + AL+EAFYR N PN+ +L+ATV VF +VIYFQGFRVD
Sbjct 205 RGTEFEGAVIALFHLLATRTDKVRALREAFYRQNLPNLMNLIATVFVFAVVIYFQGFRVD 264
Query 121 LAVKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQW 180
L +K R RGQ SYPIKLFYTSNIPIILQ+ALVSNLY +SQ+L RF N LVNLLGQW
Sbjct 265 LPIKSARYRGQYSSYPIKLFYTSNIPIILQSALVSNLYVISQMLSVRFSGNFLVNLLGQW 324
Query 181 QEVDAGG--HSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
+V GG S PVGG+ YY+SPP S G+I EDPVH VYI F+L SCA FSKTWIEVSG
Sbjct 325 ADVSGGGPARSYPVGGLCYYLSPPESMGAIFEDPVHVVVYIIFMLGSCAFFSKTWIEVSG 384
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNR 269
SS++DVAKQL++QQMVM+G+RD+S+V LNR
Sbjct 385 SSAKDVAKQLKEQQMVMRGHRDTSMVHELNR 415
> sce:YLR378C SEC61; Essential subunit of Sec61 complex (Sec61p,
Sbh1p, and Sss1p); forms a channel for SRP-dependent protein
import and retrograde transport of misfolded proteins out
of the ER; with Sec63 complex allows SRP-independent protein
import into ER; K10956 protein transport protein SEC61 subunit
alpha
Length=480
Score = 366 bits (939), Expect = 7e-101, Method: Compositional matrix adjust.
Identities = 176/315 (55%), Positives = 231/315 (73%), Gaps = 1/315 (0%)
Query 5 LLIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTE 64
LL+I QL FA ++V++LDELL KGYGLGSGISLF ATNI E I W+AF+PTT+ +G+G E
Sbjct 151 LLLIFQLMFASLIVMLLDELLSKGYGLGSGISLFTATNIAEQIFWRAFAPTTVNSGRGKE 210
Query 65 FEGALVALFHCLFTKSSNLAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVDLAVK 124
FEGA++A FH L + AL EAFYR+N PN+ +L TV +FL V+Y QGFR +L ++
Sbjct 211 FEGAVIAFFHLLAVRKDKKRALVEAFYRTNLPNMFQVLMTVAIFLFVLYLQGFRYELPIR 270
Query 125 YQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKHNVLVNLLGQWQ-EV 183
+VRGQ G YPIKLFYTSN PI+LQ+AL SN++ +SQ+L++++ N L+ L+G W
Sbjct 271 STKVRGQIGIYPIKLFYTSNTPIMLQSALTSNIFLISQILFQKYPTNPLIRLIGVWGIRP 330
Query 184 DAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSGSSSRD 243
G + + G+AYYI P S L DP+ + VYITFVL SCA+FSKTWIE+SG+S RD
Sbjct 331 GTQGPQMALSGLAYYIQPLMSLSEALLDPIKTIVYITFVLGSCAVFSKTWIEISGTSPRD 390
Query 244 VAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSGTGILL 303
+AKQ +DQ MV+ G R++S+ + L + IPTAAAFGG IGAL++ +D LG +GSG IL+
Sbjct 391 IAKQFKDQGMVINGKRETSIYRELKKIIPTAAAFGGATIGALSVGSDLLGTLGSGASILM 450
Query 304 AVTIIYQYYEMLAKE 318
A T IY YYE AKE
Sbjct 451 ATTTIYGYYEAAAKE 465
> sce:YBR283C SSH1; Ssh1p; K10956 protein transport protein SEC61
subunit alpha
Length=490
Score = 147 bits (372), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 94/323 (29%), Positives = 165/323 (51%), Gaps = 8/323 (2%)
Query 6 LIILQLFFAGVVVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKG--T 63
LI QL AG+ +L E++ KG+G SG + I +V F + IK G+ T
Sbjct 152 LINFQLVGAGIFTTLLAEVIDKGFGFSSGAMIINTVVIATNLVADTFGVSQIKVGEDDQT 211
Query 64 EFEGALVALFHCLFTKSSN-LAALKEAFYRSNAPNITSLLATVLVFLIVIYFQGFRVDLA 122
E +GAL+ L L +K + + AF R PN+T+ + + + +IV Y Q RV+L
Sbjct 212 EAQGALINLIQGLRSKHKTFIGGIISAFNRDYLPNLTTTIIVLAIAIIVCYLQSVRVELP 271
Query 123 VKYQRVRGQQGSYPIKLFYTSNIPIILQTALVSNLYF----LSQLLYRRFKHNVLVNLLG 178
++ R RG YPIKL YT + ++ ++ ++ L QL+ + +++ ++G
Sbjct 272 IRSTRARGTNNVYPIKLLYTGCLSVLFSYTILFYIHIFAFVLIQLVAKNEPTHIICKIMG 331
Query 179 QWQEVDAGGHSVPVGGIAYYISPPGSFGSILEDPVHSFVYITFVLVSCALFSKTWIEVSG 238
++ + +VP ++ P F + + P+ Y F+LV+ F+ W +SG
Sbjct 332 HYENAN-NLLAVPTFPLSLLAPPTSFFKGVTQQPLTFITYSAFILVTGIWFADKWQAISG 390
Query 239 SSSRDVAKQLRDQQMVMKGYRDSSLVQVLNRYIPTAAAFGGMCIGALTIIADFLGAIGSG 298
SS+RDVA + +DQ + + G R+ ++ + LN+ IP AA G + +T+I + LG G
Sbjct 391 SSARDVALEFKDQGITLMGRREQNVAKELNKVIPIAAVTGASVLSLITVIGESLGLKGKA 450
Query 299 TGILLAVTIIYQYYEMLAKEKEQ 321
GI++ + + E++ E +Q
Sbjct 451 AGIVVGIAGGFSLLEVITIEYQQ 473
> cel:T19D2.1 hypothetical protein
Length=872
Score = 31.2 bits (69), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query 40 ATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSSNLAAL 86
AT +C + + P TI +G+G +FE A+ ++ CL + S+N+ +
Sbjct 516 ATRVCSRLRDENAIPNTILSGEGMQFEQAMCKIW-CLISGSTNIRTV 561
> hsa:253012 HEPACAM2; HEPACAM family member 2
Length=462
Score = 30.8 bits (68), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 5/39 (12%)
Query 21 LDELLQKGYGLG-----SGISLFIATNICETIVWKAFSP 54
L++L QKG L +GISLF+ ++C +WK + P
Sbjct 339 LEKLAQKGKSLSPLASITGISLFLIISMCLLFLWKKYQP 377
Lambda K H
0.325 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13521951420
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40