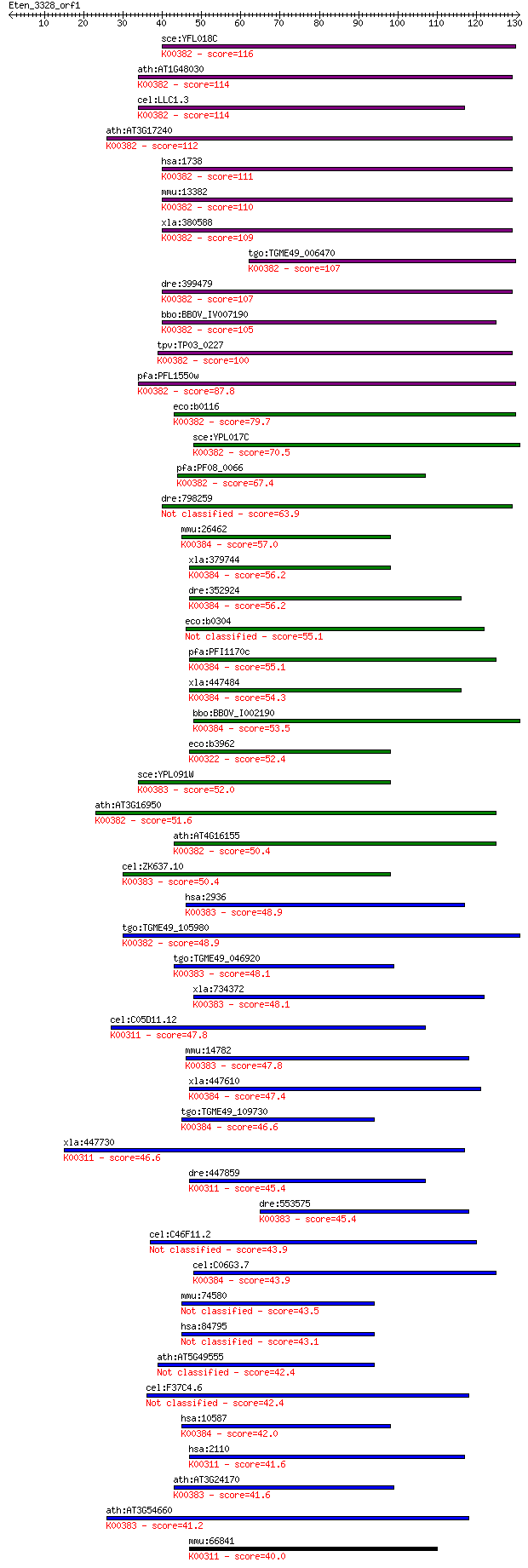
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3328_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the li... 116 1e-26
ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrog... 114 6e-26
cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehyd... 114 8e-26
ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP ... 112 3e-25
hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehy... 111 6e-25
mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogen... 110 1e-24
xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1... 109 2e-24
tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.... 107 8e-24
dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase (E... 107 9e-24
bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase (E... 105 3e-23
tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00... 100 1e-21
pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihy... 87.8 8e-18
eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydroge... 79.7 2e-15
sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogena... 70.5 1e-12
pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4); ... 67.4 1e-11
dre:798259 im:7135991; si:ch1073-179p4.3 63.9 1e-10
mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd... 57.0 2e-08
xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1... 56.2 2e-08
dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredo... 56.2 2e-08
eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide... 55.1 5e-08
pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thiore... 55.1 6e-08
xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1... 54.3 9e-08
bbo:BBOV_I002190 19.m02214; thiodoxin reductase (EC:1.8.1.9); ... 53.5 2e-07
eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotid... 52.4 4e-07
sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathio... 52.0 4e-07
ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrol... 51.6 6e-07
ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoa... 50.4 1e-06
cel:ZK637.10 trxr-2; ThioRedoXin Reductase family member (trxr... 50.4 1e-06
hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7); K0... 48.9 4e-06
tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putativ... 48.9 4e-06
tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7)... 48.1 6e-06
xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7); ... 48.1 7e-06
cel:C05D11.12 let-721; LEThal family member (let-721); K00311 ... 47.8 8e-06
mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione re... 47.8 8e-06
xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1... 47.4 1e-05
tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7)... 46.6 2e-05
xla:447730 etfdh, MGC81928; electron-transferring-flavoprotein... 46.6 2e-05
dre:447859 etfdh, wu:fa05a11, wu:fb71g06, zgc:92093; electron-... 45.4 4e-05
dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathi... 45.4 5e-05
cel:C46F11.2 hypothetical protein 43.9 1e-04
cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-... 43.9 1e-04
mmu:74580 Pyroxd2, 3830409H07Rik, 4833409A17Rik; pyridine nucl... 43.5 2e-04
hsa:84795 PYROXD2, C10orf33, FLJ12197, FLJ23849, FP3420, MGC13... 43.1 2e-04
ath:AT5G49555 amine oxidase-related 42.4 3e-04
cel:F37C4.6 hypothetical protein 42.4 4e-04
hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin r... 42.0 5e-04
hsa:2110 ETFDH, ETFQO, MADD; electron-transferring-flavoprotei... 41.6 6e-04
ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase); ... 41.6 7e-04
ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding / gl... 41.2 8e-04
mmu:66841 Etfdh, 0610010I20Rik, AV001013; electron transferrin... 40.0 0.002
> sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the
lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase
and 2-oxoglutarate dehydrogenase multi-enzyme complexes
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 116 bits (291), Expect = 1e-26, Method: Composition-based stats.
Identities = 54/91 (59%), Positives = 70/91 (76%), Gaps = 1/91 (1%)
Query 40 RSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLN 99
R+ + +DVV+IGGGP GYVAAIKAAQLG TACVEKRG LGGTCLNVGCIPSKALLN
Sbjct 19 RTLTINKSHDVVIIGGGPAGYVAAIKAAQLGFNTACVEKRGKLGGTCLNVGCIPSKALLN 78
Query 100 MSHKYKEASSDFSRFGIRVS-DVSVDISSMQ 129
SH + + ++ + GI V+ D+ +++++ Q
Sbjct 79 NSHLFHQMHTEAQKRGIDVNGDIKINVANFQ 109
> ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrogenase
1); ATP binding / dihydrolipoyl dehydrogenase; K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 114 bits (286), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 65/97 (67%), Positives = 75/97 (77%), Gaps = 2/97 (2%)
Query 34 FRRTHQRSFSSQ--SEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGC 91
F + R F+S E DVV+IGGGPGGYVAAIKA+QLGLKT C+EKRGALGGTCLNVGC
Sbjct 28 FSFSLSRGFASSGSDENDVVIIGGGPGGYVAAIKASQLGLKTTCIEKRGALGGTCLNVGC 87
Query 92 IPSKALLNMSHKYKEASSDFSRFGIRVSDVSVDISSM 128
IPSKALL+ SH Y EA F+ GI+VS V VD+ +M
Sbjct 88 IPSKALLHSSHMYHEAKHSFANHGIKVSSVEVDLPAM 124
> cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=495
Score = 114 bits (285), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 55/83 (66%), Positives = 63/83 (75%), Gaps = 0/83 (0%)
Query 34 FRRTHQRSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIP 93
F + R++S+ + D+VVIGGGPGGYVAAIKAAQLG+KT CVEK LGGTCLNVGCIP
Sbjct 16 FFQVLARNYSNTQDADLVVIGGGPGGYVAAIKAAQLGMKTVCVEKNATLGGTCLNVGCIP 75
Query 94 SKALLNMSHKYKEASSDFSRFGI 116
SKALLN SH A DF+ GI
Sbjct 76 SKALLNNSHYLHMAQHDFAARGI 98
> ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP
binding / dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=507
Score = 112 bits (280), Expect = 3e-25, Method: Composition-based stats.
Identities = 65/103 (63%), Positives = 75/103 (72%), Gaps = 0/103 (0%)
Query 26 PTVHVSALFRRTHQRSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGT 85
PT F T + S + DVV+IGGGPGGYVAAIKAAQLGLKT C+EKRGALGGT
Sbjct 22 PTDAFRFSFSLTRGFASSGSDDNDVVIIGGGPGGYVAAIKAAQLGLKTTCIEKRGALGGT 81
Query 86 CLNVGCIPSKALLNMSHKYKEASSDFSRFGIRVSDVSVDISSM 128
CLNVGCIPSKALL+ SH Y EA F+ G++VS V VD+ +M
Sbjct 82 CLNVGCIPSKALLHSSHMYHEAKHVFANHGVKVSSVEVDLPAM 124
> hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=509
Score = 111 bits (277), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 56/91 (61%), Positives = 67/91 (73%), Gaps = 2/91 (2%)
Query 40 RSFSSQS-EYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALL 98
R+++ Q + DV VIG GPGGYVAAIKAAQLG KT C+EK LGGTCLNVGCIPSKALL
Sbjct 33 RTYADQPIDADVTVIGSGPGGYVAAIKAAQLGFKTVCIEKNETLGGTCLNVGCIPSKALL 92
Query 99 NMSHKYKEA-SSDFSRFGIRVSDVSVDISSM 128
N SH Y A DF+ GI +S+V +++ M
Sbjct 93 NNSHYYHMAHGKDFASRGIEMSEVRLNLDKM 123
> mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 55/91 (60%), Positives = 66/91 (72%), Gaps = 2/91 (2%)
Query 40 RSFSSQS-EYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALL 98
R+++ Q E DV VIG GPGGYVAAIK+AQLG KT C+EK LGGTCLNVGCIPSKALL
Sbjct 33 RTYADQPIEADVTVIGSGPGGYVAAIKSAQLGFKTVCIEKNETLGGTCLNVGCIPSKALL 92
Query 99 NMSHKYKEA-SSDFSRFGIRVSDVSVDISSM 128
N SH Y A DF+ GI + +V +++ M
Sbjct 93 NNSHYYHMAHGKDFASRGIEIPEVRLNLEKM 123
> xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 55/91 (60%), Positives = 67/91 (73%), Gaps = 2/91 (2%)
Query 40 RSFSSQS-EYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALL 98
R++S ++ E DV V+G GPGGYVAAIKAAQLG +T CVEK LGGTCLNVGCIPSKALL
Sbjct 33 RNYSEKAIEADVTVVGSGPGGYVAAIKAAQLGFQTVCVEKNDTLGGTCLNVGCIPSKALL 92
Query 99 NMSHKYKEA-SSDFSRFGIRVSDVSVDISSM 128
N SH Y A DF+ GI V+ + +++ M
Sbjct 93 NNSHLYHLAHGKDFASRGIEVTGIHLNLEKM 123
> tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=519
Score = 107 bits (267), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 49/68 (72%), Positives = 57/68 (83%), Gaps = 0/68 (0%)
Query 62 AAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSHKYKEASSDFSRFGIRVSDV 121
AAIKAAQLGLKTACVEKRG LGGTCLNVGCIPSKA+LN+S+KY +A F R GI++ +
Sbjct 64 AAIKAAQLGLKTACVEKRGTLGGTCLNVGCIPSKAVLNISNKYVDARDHFERLGIKIDGL 123
Query 122 SVDISSMQ 129
S+DI MQ
Sbjct 124 SIDIDKMQ 131
> dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 107 bits (267), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 53/92 (57%), Positives = 66/92 (71%), Gaps = 3/92 (3%)
Query 40 RSFSSQS--EYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKAL 97
R+++ ++ + DV V+G GPGGYVAAIKAAQLG KT CVEK LGGTCLNVGCIPSKAL
Sbjct 31 RTYADKAAIDADVTVVGSGPGGYVAAIKAAQLGFKTVCVEKNATLGGTCLNVGCIPSKAL 90
Query 98 LNMSHKYKEA-SSDFSRFGIRVSDVSVDISSM 128
LN S+ Y A DF GI + +S+++ M
Sbjct 91 LNNSYLYHMAHGKDFESRGIEIQGISLNLEKM 122
> bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=481
Score = 105 bits (263), Expect = 3e-23, Method: Composition-based stats.
Identities = 51/86 (59%), Positives = 63/86 (73%), Gaps = 3/86 (3%)
Query 40 RSFSSQS-EYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALL 98
R FSS + +YD+ VIGGGPGGY AIKAAQ GLK AC+++R LGGTCLNVGCIPSK LL
Sbjct 15 RGFSSATHKYDLAVIGGGPGGYTTAIKAAQYGLKVACIDRRTTLGGTCLNVGCIPSKCLL 74
Query 99 NMSHKYKEASSDFSRFGIRVSDVSVD 124
N SH YK + + GI+ ++V +
Sbjct 75 NTSHHYKASHDGIA--GIKFTNVEFN 98
> tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 100 bits (249), Expect = 1e-21, Method: Composition-based stats.
Identities = 50/91 (54%), Positives = 62/91 (68%), Gaps = 3/91 (3%)
Query 39 QRSFS-SQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKAL 97
+R FS S S+YD++V+G GPGGY AIKAAQ GLK VEKR LGGTCLN GCIPSK+L
Sbjct 14 KRQFSTSSSKYDLLVLGAGPGGYTMAIKAAQHGLKVGVVEKRPTLGGTCLNCGCIPSKSL 73
Query 98 LNMSHKYKEASSDFSRFGIRVSDVSVDISSM 128
LN SH Y + G+R++ + D+ M
Sbjct 74 LNTSHLYHLMKKGVN--GLRITGLETDVGKM 102
> pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=512
Score = 87.8 bits (216), Expect = 8e-18, Method: Composition-based stats.
Identities = 50/96 (52%), Positives = 66/96 (68%), Gaps = 0/96 (0%)
Query 34 FRRTHQRSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIP 93
F + +R FS++ +YDV+VIGGGPGGYV +I+ AQ L V + LGGTCLN GCIP
Sbjct 11 FFQPLRRCFSTKKDYDVIVIGGGPGGYVCSIRCAQNKLNVLNVNEDKKLGGTCLNRGCIP 70
Query 94 SKALLNMSHKYKEASSDFSRFGIRVSDVSVDISSMQ 129
SK+LL++SH Y EA + F GI V +V +DI +M
Sbjct 71 SKSLLHISHNYYEAKTRFKECGILVDNVKLDIETMH 106
> eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydrogenase,
E3 component is part of three enzyme complexes (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=474
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 40/87 (45%), Positives = 56/87 (64%), Gaps = 1/87 (1%)
Query 43 SSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSH 102
S++ + VVV+G GP GY AA + A LGL+T VE+ LGG CLNVGCIPSKALL+++
Sbjct 2 STEIKTQVVVLGAGPAGYSAAFRCADLGLETVIVERYNTLGGVCLNVGCIPSKALLHVAK 61
Query 103 KYKEASSDFSRFGIRVSDVSVDISSMQ 129
+EA + + GI + DI ++
Sbjct 62 VIEEAKA-LAEHGIVFGEPKTDIDKIR 87
> sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=499
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 37/85 (43%), Positives = 54/85 (63%), Gaps = 2/85 (2%)
Query 48 YDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSHKYK-- 105
YDV+VIG GPGG+ AA++A+Q GL TACV++R +LGG L G +PSK LL S+ Y+
Sbjct 18 YDVLVIGCGPGGFTAAMQASQAGLLTACVDQRASLGGAYLVDGAVPSKTLLYESYLYRLL 77
Query 106 EASSDFSRFGIRVSDVSVDISSMQN 130
+ + G R+ D+ + Q+
Sbjct 78 QQQELIEQRGTRLFPAKFDMQAAQS 102
> pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=666
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 32/64 (50%), Positives = 43/64 (67%), Gaps = 1/64 (1%)
Query 44 SQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVE-KRGALGGTCLNVGCIPSKALLNMSH 102
++ EYD+ +IG G GG+ AAI A + LK +GGTC+NVGCIPSKALL ++
Sbjct 122 NEKEYDLAIIGCGVGGHAAAINAMERNLKVIIFAGDENCIGGTCVNVGCIPSKALLYATN 181
Query 103 KYKE 106
KY+E
Sbjct 182 KYRE 185
> dre:798259 im:7135991; si:ch1073-179p4.3
Length=371
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 38/99 (38%), Positives = 60/99 (60%), Gaps = 11/99 (11%)
Query 40 RSFSS-QSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVG 90
R+F++ + +YD+VVIGGG GG + +AAQLG K A ++ + LGGTC+NVG
Sbjct 24 RNFTAGKFDYDLVVIGGGSGGLACSKEAAQLGQKVAVLDYVEPSLKGTKWGLGGTCVNVG 83
Query 91 CIPSKALLNMSHKYKEASSDFSRFGIRVSD-VSVDISSM 128
CIP K L++ + A D ++G ++ + +S D +M
Sbjct 84 CIPKK-LMHQAALLGTAVKDARKYGWQIPETLSHDWPTM 121
> mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd2;
thioredoxin reductase 2 (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=527
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 37/61 (60%), Gaps = 8/61 (13%)
Query 45 QSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIPSKA 96
Q +D++VIGGG GG A +AAQLG K A + + LGGTC+NVGCIP K
Sbjct 40 QQSFDLLVIGGGSGGLACAKEAAQLGKKVAVADYVEPSPRGTKWGLGGTCVNVGCIPKKL 99
Query 97 L 97
+
Sbjct 100 M 100
> xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=504
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 29/59 (49%), Positives = 37/59 (62%), Gaps = 8/59 (13%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLGLKTACVE-----KRG---ALGGTCLNVGCIPSKAL 97
+YD++VIGGG GG A +AAQ G K A + RG +GGTC+NVGCIP K +
Sbjct 19 DYDLLVIGGGSGGLACAKQAAQFGKKVAVFDYVEPSPRGTKWGIGGTCVNVGCIPKKLM 77
> dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=602
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 43/77 (55%), Gaps = 9/77 (11%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIPSKALL 98
+YD++VIGGG GG + +AA LG K ++ LGGTC+NVGCIP K L+
Sbjct 115 DYDLIVIGGGSGGLACSKEAATLGKKVMVLDYVVPTPQGTAWGLGGTCVNVGCIPKK-LM 173
Query 99 NMSHKYKEASSDFSRFG 115
+ + A D +FG
Sbjct 174 HQTALLGTAMEDARKFG 190
> eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide-disulfide
oxidoreductase
Length=441
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 48/77 (62%), Gaps = 5/77 (6%)
Query 46 SEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGAL-GGTCLNVGCIPSKALLNMSHKY 104
++Y V+IG G G A+ A+ G + A +E+ A+ GGTC+N+GCIP+K L++ + ++
Sbjct 2 NKYQAVIIGFGKAGKTLAVTLAKAGWRVALIEQSNAMYGGTCINIGCIPTKTLVHDAQQH 61
Query 105 KEASSDFSRFGIRVSDV 121
+DF R R ++V
Sbjct 62 ----TDFVRAIQRKNEV 74
> pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=617
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 30/86 (34%), Positives = 44/86 (51%), Gaps = 8/86 (9%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLG--------LKTACVEKRGALGGTCLNVGCIPSKALL 98
+YD VVIGGGPGG +A +AA G +K + + +GGTC+NVGC+P K +
Sbjct 117 DYDYVVIGGGPGGMASAKEAAAHGARVLLFDYVKPSSQGTKWGIGGTCVNVGCVPKKLMH 176
Query 99 NMSHKYKEASSDFSRFGIRVSDVSVD 124
H D +G + ++ D
Sbjct 177 YAGHMGSIFKLDSKAYGWKFDNLKHD 202
> xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=596
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 9/77 (11%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIPSKALL 98
+YD++VIGGG GG + +AA G K ++ LGGTC+NVGCIP K L+
Sbjct 111 DYDLIVIGGGSGGLACSKEAASFGKKVMVLDFVVPTPQGTSWGLGGTCVNVGCIPKK-LM 169
Query 99 NMSHKYKEASSDFSRFG 115
+ + ++ D +FG
Sbjct 170 HQAAILGQSLKDSRKFG 186
> bbo:BBOV_I002190 19.m02214; thiodoxin reductase (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=559
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/95 (38%), Positives = 49/95 (51%), Gaps = 13/95 (13%)
Query 48 YDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIPSKALLN 99
YD+ VIGGG G AA +AA+LG KT + + LGGTC+NVGCIP K L++
Sbjct 69 YDLAVIGGGCSGLAAAKEAARLGAKTVLFDYVRPSPRGTKWGLGGTCVNVGCIPKK-LMH 127
Query 100 MSHKYKEASSDFSRFGIRVS----DVSVDISSMQN 130
+ A D G + D S I ++QN
Sbjct 128 YAGILGHAEHDREMLGWSDASPKHDWSKMIQTIQN 162
> eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotide
transhydrogenase, soluble (EC:1.6.1.1); K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=466
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/51 (47%), Positives = 33/51 (64%), Gaps = 0/51 (0%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKAL 97
+YD +VIG GPGG AA+ + G + A +E+ +GG C + G IPSKAL
Sbjct 6 DYDAIVIGSGPGGEGAAMGLVKQGARVAVIERYQNVGGGCTHWGTIPSKAL 56
> sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathione
oxidoreductase, converts oxidized glutathione to reduced
glutathione; mitochondrial but not cytosolic form has a role
in resistance to hyperoxia (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 33/65 (50%), Positives = 44/65 (67%), Gaps = 2/65 (3%)
Query 34 FRRTHQRSFSSQSE-YDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCI 92
FR R+ S+ ++ YD +VIGGG GG +A +AA G KT VE + ALGGTC+NVGC+
Sbjct 9 FRSLQIRTMSTNTKHYDYLVIGGGSGGVASARRAASYGAKTLLVEAK-ALGGTCVNVGCV 67
Query 93 PSKAL 97
P K +
Sbjct 68 PKKVM 72
> ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrolipoyl
dehydrogenase; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=623
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 43/114 (37%), Positives = 67/114 (58%), Gaps = 13/114 (11%)
Query 23 LTVPTVHVSALFRRTHQRSFSSQS----------EYDVVVIGGGPGGYVAAIKAAQLGLK 72
L + + + L R++ Q S S+ S +YD+++IG G GG+ AA+ A + GLK
Sbjct 52 LAIRSNRIQFLSRKSFQVSASASSNGNGAPPKSFDYDLIIIGAGVGGHGAALHAVEKGLK 111
Query 73 TACVEKRGALGGTCLNVGCIPSKALLNMSHKYKEASSD--FSRFGIRVSDVSVD 124
TA +E +GGTC+N GC+PSKALL +S + +E ++ FG++VS D
Sbjct 112 TAIIEGD-VVGGTCVNRGCVPSKALLAVSGRMRELQNEHHMKSFGLQVSAAGYD 164
> ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=630
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/84 (44%), Positives = 55/84 (65%), Gaps = 3/84 (3%)
Query 43 SSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSH 102
S +YD+++IG G GG+ AA+ A + GLKTA +E +GGTC+N GC+PSKALL +S
Sbjct 143 SKSFDYDLIIIGAGVGGHGAALHAVEKGLKTAIIEGD-VVGGTCVNRGCVPSKALLAVSG 201
Query 103 KYKEASSD--FSRFGIRVSDVSVD 124
+ +E ++ FG++VS D
Sbjct 202 RMRELQNEHHMKAFGLQVSAAGYD 225
> cel:ZK637.10 trxr-2; ThioRedoXin Reductase family member (trxr-2);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=503
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 39/76 (51%), Gaps = 8/76 (10%)
Query 30 VSALFRRTHQRSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEK--------RGA 81
+S R R S +++D++VIG G GG + +AA LG A ++
Sbjct 3 LSTFKRHLPIRRLFSSNKFDLIVIGAGSGGLSCSKRAADLGANVALIDAVEPTPHGHSWG 62
Query 82 LGGTCLNVGCIPSKAL 97
+GGTC NVGCIP K +
Sbjct 63 IGGTCANVGCIPKKLM 78
> hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=522
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 45/71 (63%), Gaps = 2/71 (2%)
Query 46 SEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSHKYK 105
+ YD +VIGGG GG +A +AA+LG + A VE LGGTC+NVGC+P K + N + +
Sbjct 63 ASYDYLVIGGGSGGLASARRAAELGARAAVVESH-KLGGTCVNVGCVPKKVMWNTA-VHS 120
Query 106 EASSDFSRFGI 116
E D + +G
Sbjct 121 EFMHDHADYGF 131
> tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putative
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=636
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 45/104 (43%), Positives = 63/104 (60%), Gaps = 5/104 (4%)
Query 30 VSALFRRTHQRSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNV 89
VS LF + +FS + +DV +IG G GG+ AA+ AA LGLKTA V G GGTC+N
Sbjct 125 VSRLFATSSSTNFSDEP-FDVTIIGLGVGGHAAALHAAALGLKTAVVSG-GDPGGTCVNR 182
Query 90 GCIPSKALLNMSHKYKEASSD--FSRFGIRVS-DVSVDISSMQN 130
GC+PSKALL + + K + S G++V ++ VD + + N
Sbjct 183 GCVPSKALLAAARRVKMLRNKHHLSAMGLQVEGEIKVDPTGVGN 226
> tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 34/56 (60%), Gaps = 1/56 (1%)
Query 43 SSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALL 98
+++ +D+ VIGGG GG A +AA ++ + LGGTC+NVGC+P K +
Sbjct 8 ATRRHFDLFVIGGGSGGLACARRAATYNVRVGLADGN-RLGGTCVNVGCVPKKVMW 62
> xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=476
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 48/74 (64%), Gaps = 2/74 (2%)
Query 48 YDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSHKYKEA 107
YD +V+GGG GG +A +AA+LG +TA VE LGGTC+NVGC+P K + N + + E
Sbjct 19 YDYLVVGGGSGGLASARRAAELGARTAVVES-SKLGGTCVNVGCVPKKIMWNAAM-HSEY 76
Query 108 SSDFSRFGIRVSDV 121
D + +G + DV
Sbjct 77 IHDHADYGFEIPDV 90
> cel:C05D11.12 let-721; LEThal family member (let-721); K00311
electron-transferring-flavoprotein dehydrogenase [EC:1.5.5.1]
Length=597
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 32/91 (35%), Positives = 46/91 (50%), Gaps = 11/91 (12%)
Query 27 TVHVSALFRRTHQR----SFSSQSE-YDVVVIGGGPGGYVAAIKAAQLG------LKTAC 75
T H + R T R + +S+ YDVV++GGGP G AAI+ QL L+
Sbjct 25 TTHYTVKDRSTDPRWKDVDLARESDVYDVVIVGGGPSGLSAAIRLRQLAEKAQKELRVCV 84
Query 76 VEKRGALGGTCLNVGCIPSKALLNMSHKYKE 106
VEK +GG L+ I ++AL + +KE
Sbjct 85 VEKASVIGGHTLSGAVIETRALDELIPNWKE 115
> mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione
reductase (EC:1.8.1.7); K00383 glutathione reductase (NADPH)
[EC:1.8.1.7]
Length=500
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 46 SEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSHKYK 105
S +D +VIGGG GG +A +AA+LG + A VE LGGTC+NVGC+P K + N + +
Sbjct 41 SSFDYLVIGGGSGGLASARRAAELGARAAVVESH-KLGGTCVNVGCVPKKVMWNTA-VHS 98
Query 106 EASSDFSRFGIR 117
E D +G +
Sbjct 99 EFMHDHVDYGFQ 110
> xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=531
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 31/82 (37%), Positives = 51/82 (62%), Gaps = 9/82 (10%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIPSKALL 98
+YD++VIGGG GG A+ +AA+ G K ++ + LGGTC+NVGCIP K L+
Sbjct 47 DYDLIVIGGGSGGLAASKEAAKYGKKVLVLDFVTPSPLGTKWGLGGTCVNVGCIPKK-LM 105
Query 99 NMSHKYKEASSDFSRFGIRVSD 120
+ + ++A D ++G +++D
Sbjct 106 HQAALLRQALKDSQKYGWQIAD 127
> tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=662
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/57 (42%), Positives = 31/57 (54%), Gaps = 8/57 (14%)
Query 45 QSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIP 93
+ +YD+ VIGGG GG A AA G +T + LGGTC+NVGC+P
Sbjct 157 EFDYDLAVIGGGSGGLACAKMAAAQGAETVVFDFVQPSTQGSTWGLGGTCVNVGCVP 213
> xla:447730 etfdh, MGC81928; electron-transferring-flavoprotein
dehydrogenase (EC:1.5.5.1); K00311 electron-transferring-flavoprotein
dehydrogenase [EC:1.5.5.1]
Length=616
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 37/121 (30%), Positives = 54/121 (44%), Gaps = 21/121 (17%)
Query 15 LHRLAARPLTVPTVHVSALFRRTHQR-------SFSSQSEYDVVVIGGGPGGYVAAIKAA 67
L R A+ + T H + R QR F+ E DVV++GGGP G AAI+
Sbjct 30 LKRWASSSVPRITTHYTIYPRGQDQRWEGVNMERFAE--EADVVIVGGGPAGLSAAIRLK 87
Query 68 QLG------LKTACVEKRGALGGTCLNVGCIPSKALLNMSHKYKEASSDFS------RFG 115
QL L+ VEK +G L+ C+ +AL + +KE + + +FG
Sbjct 88 QLAAECDKELRVCLVEKGAQIGAHILSGACLEPRALEELFPDWKERGAPLNTPVTEDKFG 147
Query 116 I 116
I
Sbjct 148 I 148
> dre:447859 etfdh, wu:fa05a11, wu:fb71g06, zgc:92093; electron-transferring-flavoprotein
dehydrogenase (EC:1.5.5.1); K00311
electron-transferring-flavoprotein dehydrogenase [EC:1.5.5.1]
Length=617
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLG------LKTACVEKRGALGGTCLNVGCIPSKALLNM 100
E DVV+IGGGP G AAI+ QL L+ VEK +G L+ C+ AL +
Sbjct 68 EADVVIIGGGPAGLSAAIRLKQLANQHEKELRVCVVEKASQIGAHTLSGACLEPTALNEL 127
Query 101 SHKYKE 106
+KE
Sbjct 128 IPDWKE 133
> dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=425
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 31/53 (58%), Gaps = 2/53 (3%)
Query 65 KAAQLGLKTACVEKRGALGGTCLNVGCIPSKALLNMSHKYKEASSDFSRFGIR 117
+AA+LG TA +E LGGTC+NVGC+P K + N S + E D +G
Sbjct 26 RAAELGATTAVIESH-RLGGTCVNVGCVPKKVMWNTS-THAEYLHDHEDYGFE 76
> cel:C46F11.2 hypothetical protein
Length=473
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 33/83 (39%), Positives = 48/83 (57%), Gaps = 2/83 (2%)
Query 37 THQRSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIPSKA 96
T + S E+D +VIGGG GG +A +A + G+ +E G LGGTC+NVGC+P K
Sbjct 10 TSRSIMSGVKEFDYLVIGGGSGGIASARRAREFGVSVGLIES-GRLGGTCVNVGCVPKKV 68
Query 97 LLNMSHKYKEASSDFSRFGIRVS 119
+ N S + E D + +G V+
Sbjct 69 MYNCS-LHAEFIRDHADYGFDVT 90
> cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-1);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=667
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 33/85 (38%), Positives = 51/85 (60%), Gaps = 9/85 (10%)
Query 48 YDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIPSKALLN 99
YD++VIGGG GG AA +A++LG K AC++ LGGTC+NVGCIP K L++
Sbjct 173 YDLIVIGGGSGGLAAAKEASRLGKKVACLDFVKPSPQGTSWGLGGTCVNVGCIPKK-LMH 231
Query 100 MSHKYKEASSDFSRFGIRVSDVSVD 124
+ + D ++G ++ + V+
Sbjct 232 QASLLGHSIHDAKKYGWKLPEGKVE 256
> mmu:74580 Pyroxd2, 3830409H07Rik, 4833409A17Rik; pyridine nucleotide-disulphide
oxidoreductase domain 2
Length=581
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 45 QSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIP 93
+ EYD VVIG G G VAA +LG+ TA E+R +GG + IP
Sbjct 32 KPEYDAVVIGAGHNGLVAAAYLQRLGVNTAVFERRHVIGGAAVTEEIIP 80
> hsa:84795 PYROXD2, C10orf33, FLJ12197, FLJ23849, FP3420, MGC13047;
pyridine nucleotide-disulphide oxidoreductase domain
2
Length=581
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 45 QSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIP 93
+ EYD VVIG G G VAA +LG+ TA E+R +GG + IP
Sbjct 32 KPEYDAVVIGAGHNGLVAAAYLQRLGVNTAVFERRHVIGGAAVTEEIIP 80
> ath:AT5G49555 amine oxidase-related
Length=556
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 34/57 (59%), Gaps = 2/57 (3%)
Query 39 QRSFSS--QSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIP 93
+RSFS+ + ++D VVIGGG G AA A+ GL A +E+R +GG + +P
Sbjct 3 RRSFSTLPKKKWDAVVIGGGHNGLTAAAYLARGGLSVAVLERRHVIGGAAVTEEIVP 59
> cel:F37C4.6 hypothetical protein
Length=544
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 42/88 (47%), Gaps = 7/88 (7%)
Query 36 RTHQRSFSSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVEKRGALGGTCLNVGCIP-- 93
R +R FS QS YD ++IGGG G AA + G K +E+R +GG + +P
Sbjct 4 RIGRRCFSKQS-YDAIIIGGGHNGLTAAAYLTKAGKKVCVLERRHVVGGAAVTEEIVPGF 62
Query 94 ----SKALLNMSHKYKEASSDFSRFGIR 117
+ LL++ + +FG+R
Sbjct 63 RFSRASYLLSLLRPVVMQELNLKKFGLR 90
> hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin
reductase 2 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH)
[EC:1.8.1.9]
Length=524
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 39/61 (63%), Gaps = 8/61 (13%)
Query 45 QSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVE--------KRGALGGTCLNVGCIPSKA 96
Q +YD++V+GGG GG A +AAQLG K A V+ R LGGTC+NVGCIP K
Sbjct 37 QRDYDLLVVGGGSGGLACAKEAAQLGRKVAVVDYVEPSPQGTRWGLGGTCVNVGCIPKKL 96
Query 97 L 97
+
Sbjct 97 M 97
> hsa:2110 ETFDH, ETFQO, MADD; electron-transferring-flavoprotein
dehydrogenase (EC:1.5.5.1); K00311 electron-transferring-flavoprotein
dehydrogenase [EC:1.5.5.1]
Length=617
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 25/82 (30%), Positives = 38/82 (46%), Gaps = 12/82 (14%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLG------LKTACVEKRGALGGTCLNVGCIPSKALLNM 100
E DVV++G GP G AA++ QL ++ VEK +G L+ C+ A +
Sbjct 68 EADVVIVGAGPAGLSAAVRLKQLAVAHEKDIRVCLVEKAAQIGAHTLSGACLDPGAFKEL 127
Query 101 SHKYKEASSDFS------RFGI 116
+KE + + RFGI
Sbjct 128 FPDWKEKGAPLNTPVTEDRFGI 149
> ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase);
FAD binding / NADP or NADPH binding / glutathione-disulfide
reductase/ oxidoreductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=499
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 34/65 (52%), Gaps = 9/65 (13%)
Query 43 SSQSEYDVVVIGGGPGGYVAAIKAAQLGLKTACVE---------KRGALGGTCLNVGCIP 93
++ ++D+ VIG G GG AA +A G K E + G +GGTC+ GC+P
Sbjct 21 ATHYDFDLFVIGAGSGGVRAARFSANHGAKVGICELPFHPISSEEIGGVGGTCVIRGCVP 80
Query 94 SKALL 98
K L+
Sbjct 81 KKILV 85
> ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding /
glutathione-disulfide reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=565
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 45/113 (39%), Gaps = 22/113 (19%)
Query 26 PTVHVSALFRRTHQRSFS------------SQSEYDVVVIGGGPGGYVAAIKAAQLGLKT 73
P + + + R H R FS ++D+ IG G GG A+ A G
Sbjct 54 PRIALLSNHRYYHSRRFSVCASTDNGAESDRHYDFDLFTIGAGSGGVRASRFATSFGASA 113
Query 74 ACVE---------KRGALGGTCLNVGCIPSKALLNMSHKYKEASSDFSRFGIR 117
A E G +GGTC+ GC+P K LL + KY D FG +
Sbjct 114 AVCELPFSTISSDTAGGVGGTCVLRGCVPKK-LLVYASKYSHEFEDSHGFGWK 165
> mmu:66841 Etfdh, 0610010I20Rik, AV001013; electron transferring
flavoprotein, dehydrogenase (EC:1.5.5.1); K00311 electron-transferring-flavoprotein
dehydrogenase [EC:1.5.5.1]
Length=616
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 33/69 (47%), Gaps = 6/69 (8%)
Query 47 EYDVVVIGGGPGGYVAAIKAAQLG------LKTACVEKRGALGGTCLNVGCIPSKALLNM 100
E DVV++G GP G AAI+ QL ++ VEK +G L+ C+ A +
Sbjct 67 EADVVIVGAGPAGLSAAIRLKQLAAEQGKDIRVCLVEKAAQIGAHTLSGACLDPAAFKEL 126
Query 101 SHKYKEASS 109
+KE +
Sbjct 127 FPDWKEKGA 135
Lambda K H
0.321 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40