bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3307_orf6
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
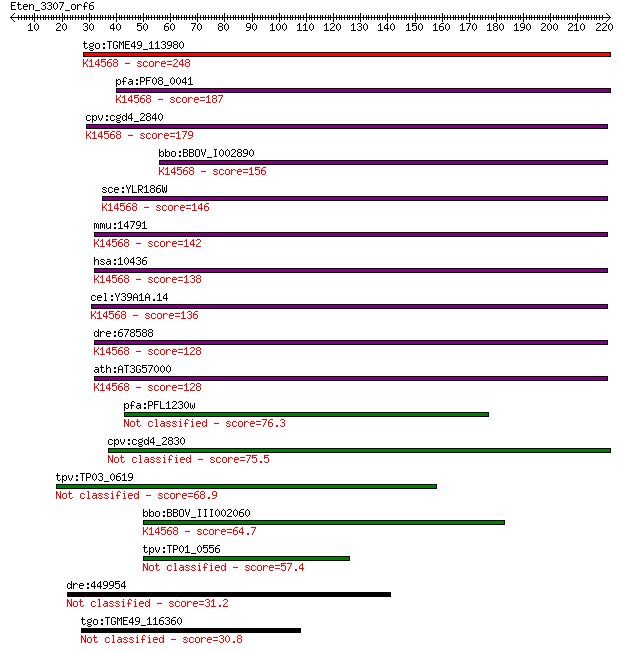
tgo:TGME49_113980 nucleolar essential protein 1, putative ; K1... 248 1e-65
pfa:PF08_0041 ribosome biogenesis protein nep1 homologue, puta... 187 3e-47
cpv:cgd4_2840 Mra1/NEP1 like protein, involved in pre-rRNA pro... 179 1e-44
bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain cont... 156 5e-38
sce:YLR186W EMG1, NEP1; Emg1p; K14568 essential for mitotic gr... 146 6e-35
mmu:14791 Emg1, C2f, Grcc2f; EMG1 nucleolar protein homolog (S... 142 7e-34
hsa:10436 EMG1, C2F, FLJ60792, Grcc2f, NEP1; EMG1 nucleolar pr... 138 1e-32
cel:Y39A1A.14 hypothetical protein; K14568 essential for mitot... 136 4e-32
dre:678588 MGC136360; zgc:136360; K14568 essential for mitotic... 128 1e-29
ath:AT3G57000 nucleolar essential protein-related; K14568 esse... 128 2e-29
pfa:PFL1230w conserved Plasmodium protein 76.3 8e-14
cpv:cgd4_2830 Mra1/NEP1 like protein, involved in pre-rRNA pro... 75.5 1e-13
tpv:TP03_0619 hypothetical protein 68.9 1e-11
bbo:BBOV_III002060 17.m07199; hypothetical protein; K14568 ess... 64.7 2e-10
tpv:TP01_0556 hypothetical protein 57.4 4e-08
dre:449954 im:7141282; si:ch73-37h15.2 31.2 3.2
tgo:TGME49_116360 hypothetical protein 30.8 3.8
> tgo:TGME49_113980 nucleolar essential protein 1, putative ;
K14568 essential for mitotic growth 1
Length=235
Score = 248 bits (634), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 117/194 (60%), Positives = 150/194 (77%), Gaps = 0/194 (0%)
Query 28 GREGSLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSG 87
G++ SLQLL +H++LL++ +R +VRPDI H CL++LQESPLN+AGRLCVFIRT
Sbjct 42 GKDRSLQLLNSLEHKQLLRKCDRQGDEVRPDIAHHCLLSLQESPLNRAGRLCVFIRTADR 101
Query 88 QLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIA 147
QLIE+ P L VPPTY EF KLM NLL+ RR++AVEKNVTL Q+ KN+ FLP S K+A
Sbjct 102 QLIEISPLLTVPPTYQEFAKLMTNLLYARRLKAVEKNVTLAQIVKNEHANFLPPNSVKVA 161
Query 148 LSTQGRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCC 207
L+ GR V L DFVQ+F++T PV+F+VGAVAH++PT + ++ ISIA GL+AAVCC
Sbjct 162 LTVSGRSVALSDFVQRFKETDTPVVFVVGAVAHSDPTGECDYVDDKISIAGVGLTAAVCC 221
Query 208 SSICAEFENLWNIY 221
SSICAEFE LW+I+
Sbjct 222 SSICAEFEALWDIF 235
> pfa:PF08_0041 ribosome biogenesis protein nep1 homologue, putative;
K14568 essential for mitotic growth 1
Length=279
Score = 187 bits (474), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 81/185 (43%), Positives = 127/185 (68%), Gaps = 3/185 (1%)
Query 40 QHRKLLQQ---ENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQL 96
+H+ +L++ + +I + RPDI HQCLI L ESPLNK G L ++I+T QL V L
Sbjct 95 KHKNILKKNKVDEENIKNFRPDILHQCLIHLLESPLNKFGYLQIYIKTHDNQLFYVSSNL 154
Query 97 RVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRPVD 156
++P T+ +F LM+ L +I+A EKN+ L+++ KND LP+ KI LS +G+ V+
Sbjct 155 KIPKTFQQFESLMVTFLRKYKIKANEKNIYLLKIIKNDLQNILPINGHKIGLSLKGKKVE 214
Query 157 LGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSICAEFEN 216
L ++++ +++T PV F +GAVA++NPT ++ ++ ISI+ LSAA+CCSSIC+EFE+
Sbjct 215 LNNYIKVYKNTNQPVTFFIGAVAYSNPTMKLQILDDNISISDFSLSAAMCCSSICSEFEH 274
Query 217 LWNIY 221
LWN++
Sbjct 275 LWNLF 279
> cpv:cgd4_2840 Mra1/NEP1 like protein, involved in pre-rRNA processing,
adjacent genes paralogs ; K14568 essential for mitotic
growth 1
Length=223
Score = 179 bits (453), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 94/210 (44%), Positives = 129/210 (61%), Gaps = 18/210 (8%)
Query 29 REGSLQLLEGFQHRKLLQQENRS------------IADVRPDITHQCLIALQESPLNKAG 76
++ SL+LL G H K +E S I ++RPDI HQCL++L +SPLNK+G
Sbjct 13 KDKSLELLNGIDHPKKKIEEYCSLYSSKIFSKEEMILNIRPDILHQCLLSLLDSPLNKSG 72
Query 77 RLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDAN 136
RL V+IRT S LIEV+PQL VP ++ EF LM+NLL R+I AV N +LM++ KND +
Sbjct 73 RLLVYIRTMSNVLIEVNPQLSVPRSFKEFSSLMVNLLVKRKITAVNGNTSLMRIVKNDID 132
Query 137 LFLPVGSRKIALSTQGRPVDLGDFVQQFQDTKN------PVLFLVGAVAHANPTTNSELA 190
LPVG +K LS G ++ + + +++ V F+VGAVA+ +P + +
Sbjct 133 KILPVGGKKYGLSLNGTQKNIRALINELYSSEDYKIRNSSVTFVVGAVAYGDPIQSCDFI 192
Query 191 EECISIAPCGLSAAVCCSSICAEFENLWNI 220
EE ISI+ LSAA+CCS IC EFE LW I
Sbjct 193 EEIISISSYPLSAALCCSKICNEFEYLWKI 222
> bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain containing
protein; K14568 essential for mitotic growth 1
Length=188
Score = 156 bits (395), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 76/165 (46%), Positives = 104/165 (63%), Gaps = 0/165 (0%)
Query 56 RPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHT 115
R DI HQ L+ L +SPLNK G L V IRT G L+EV P LRVP T +F L++ LL+
Sbjct 23 RVDILHQSLLVLLDSPLNKNGFLKVLIRTDDGNLVEVSPHLRVPRTPKQFESLLVYLLYK 82
Query 116 RRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRPVDLGDFVQQFQDTKNPVLFLV 175
R +++ E++ LM+ KN+ + LP GSR+ LS GR V L DF QF+ PV+F +
Sbjct 83 RHVKSQERDAILMRFIKNEIEIALPPGSRRFGLSVGGRQVKLKDFCHQFKAVDYPVVFHI 142
Query 176 GAVAHANPTTNSELAEECISIAPCGLSAAVCCSSICAEFENLWNI 220
GAV+H +P E EE +SI+ GL+AA C+ +C+EFE L +
Sbjct 143 GAVSHTHPKGTVEHVEEVLSISDHGLTAAHVCAKVCSEFEYLLGV 187
> sce:YLR186W EMG1, NEP1; Emg1p; K14568 essential for mitotic
growth 1
Length=252
Score = 146 bits (368), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 75/186 (40%), Positives = 111/186 (59%), Gaps = 1/186 (0%)
Query 35 LLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHP 94
LL H+ LL++ R I++ RPDITHQCL+ L +SP+NKAG+L V+I+T G LIEV+P
Sbjct 67 LLNCDDHQGLLKKMGRDISEARPDITHQCLLTLLDSPINKAGKLQVYIQTSRGILIEVNP 126
Query 95 QLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRP 154
+R+P T+ F LM+ LLH IR+V L++V KN LP RK+ LS
Sbjct 127 TVRIPRTFKRFSGLMVQLLHKLSIRSVNSEEKLLKVIKNPITDHLPTKCRKVTLSFDAPV 186
Query 155 VDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSICAEF 214
+ + D++++ D ++ +F VGA+A E +E + ++ LSA+V CS C
Sbjct 187 IRVQDYIEKLDDDESICVF-VGAMARGKDNFADEYVDEKVGLSNYPLSASVACSKFCHGA 245
Query 215 ENLWNI 220
E+ WNI
Sbjct 246 EDAWNI 251
> mmu:14791 Emg1, C2f, Grcc2f; EMG1 nucleolar protein homolog
(S. cerevisiae); K14568 essential for mitotic growth 1
Length=244
Score = 142 bits (359), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 76/189 (40%), Positives = 113/189 (59%), Gaps = 5/189 (2%)
Query 32 SLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIE 91
+ +LL +H+ +L + R +VRPDITHQ L+ L +SPLN+AG L V+I TQ LIE
Sbjct 60 TYELLNCDRHKSMLLKNGRDPGEVRPDITHQSLLMLMDSPLNRAGLLQVYIHTQKNVLIE 119
Query 92 VHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQ 151
V+PQ R+P T+D F LM+ LLH +RA + L++V KN + PVG KI S
Sbjct 120 VNPQTRIPRTFDRFCGLMVQLLHKLSVRAADGPQKLLKVIKNPVSDHFPVGCMKIGTSFS 179
Query 152 GRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSIC 211
D+ D +++ + +PV+F+VGA AH + E E+ +SI+ LSAA+ C+ +
Sbjct 180 VE--DISD-IRELVPSSDPVVFVVGAFAHGK--VSVEYTEKMVSISNYPLSAALTCAKVT 234
Query 212 AEFENLWNI 220
FE +W +
Sbjct 235 TAFEEVWGV 243
> hsa:10436 EMG1, C2F, FLJ60792, Grcc2f, NEP1; EMG1 nucleolar
protein homolog (S. cerevisiae); K14568 essential for mitotic
growth 1
Length=244
Score = 138 bits (348), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 72/189 (38%), Positives = 111/189 (58%), Gaps = 5/189 (2%)
Query 32 SLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIE 91
+ +LL +H+ +L + R + RPDITHQ L+ L +SPLN+AG L V+I TQ LIE
Sbjct 60 TYELLNCDKHKSILLKNGRDPGEARPDITHQSLLMLMDSPLNRAGLLQVYIHTQKNVLIE 119
Query 92 VHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQ 151
V+PQ R+P T+D F LM+ LLH +RA + L++V KN + PVG K+ S
Sbjct 120 VNPQTRIPRTFDRFCGLMVQLLHKLSVRAADGPQKLLKVIKNPVSDHFPVGCMKVGTSFS 179
Query 152 GRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSIC 211
+ + V++ + +P++F+VGA AH + E E+ +SI+ LSAA+ C+ +
Sbjct 180 ---IPVVSDVRELVPSSDPIVFVVGAFAHGK--VSVEYTEKMVSISNYPLSAALTCAKLT 234
Query 212 AEFENLWNI 220
FE +W +
Sbjct 235 TAFEEVWGV 243
> cel:Y39A1A.14 hypothetical protein; K14568 essential for mitotic
growth 1
Length=231
Score = 136 bits (343), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 73/190 (38%), Positives = 108/190 (56%), Gaps = 3/190 (1%)
Query 31 GSLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLI 90
G +L +H L+++ + AD RPDI HQCL+ L +SPLN+AG+L VF RT L+
Sbjct 44 GEYAILSSDKHANFLRKQKKDPADYRPDILHQCLLNLLDSPLNRAGKLRVFFRTSKNVLV 103
Query 91 EVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALST 150
+V PQ R+P T+D F LM+ LLH IRA E LM V KN + LPVGSRK+ +S
Sbjct 104 DVSPQCRIPRTFDRFCGLMVQLLHKLSIRAAETTQKLMSVVKNPVSNHLPVGSRKMLMSF 163
Query 151 QGRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSSI 210
+ + + + +T P++ ++G +A + +E IS P LSAA+ C+ +
Sbjct 164 NVPELTMANKLVA-PETDEPLVLIIGGIARGKIVVDYNDSETKISNYP--LSAALTCAKV 220
Query 211 CAEFENLWNI 220
+ E +W I
Sbjct 221 TSGLEEIWGI 230
> dre:678588 MGC136360; zgc:136360; K14568 essential for mitotic
growth 1
Length=238
Score = 128 bits (322), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 69/191 (36%), Positives = 107/191 (56%), Gaps = 9/191 (4%)
Query 32 SLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIE 91
+ +LL QH+ ++ + R +RPDI HQCL+ L +SPLN+AG L V+I T+ LIE
Sbjct 54 TFELLNCDQHKSMIIKSGRDPGKIRPDIAHQCLLMLLDSPLNRAGLLQVYIHTEKNVLIE 113
Query 92 VHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQ 151
++PQ R+P T+ F LM+ LLH +RA + L+++ KN + LP G + + S +
Sbjct 114 INPQTRIPRTFARFCGLMVQLLHKLSVRAADGPQRLLRLIKNPVSDHLPPGCPRFSTSFK 173
Query 152 GRPVDLGDFV--QQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCSS 209
GD V + P ++GA AH N + E+ +SI+ LSAA+ C+
Sbjct 174 A-----GDAVCPRTIVPDDGPAAIVIGAFAHG--AVNVDYTEKTVSISNYPLSAALACAK 226
Query 210 ICAEFENLWNI 220
IC+ FE +W +
Sbjct 227 ICSAFEEVWGV 237
> ath:AT3G57000 nucleolar essential protein-related; K14568 essential
for mitotic growth 1
Length=298
Score = 128 bits (321), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 74/192 (38%), Positives = 105/192 (54%), Gaps = 5/192 (2%)
Query 32 SLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRL-CVFIRTQSGQLI 90
+ QLL H L++ NR+ AD RPDITHQ L+ + +SP+NKAGRL V++RT+ G L
Sbjct 108 TYQLLNSDDHANFLKKNNRNPADYRPDITHQALLMILDSPVNKAGRLKAVYVRTEKGVLF 167
Query 91 EVHPQLRVPPTYDEFRKLMINLLHTRRIRAVEKNVTLMQVTKND-ANLFLPVGSRKIALS 149
EV P +R+P T+ F +M+ LL I AV L++ KN LPV S +I S
Sbjct 168 EVKPHVRIPRTFKRFAGIMLQLLQKLSITAVNSREKLLRCVKNPIEEHHLPVNSHRIGFS 227
Query 150 -TQGRPVDLGDFVQQFQDTKNPVLFLVGAVAHANPTTNSELAEECISIAPCGLSAAVCCS 208
+ + V++ + D +F+VGA+AH N +E +S++ LSAA C S
Sbjct 228 HSSEKLVNMQKHLATVCDDDRDTVFVVGAMAHGKIDCN--YIDEFVSVSEYPLSAAYCIS 285
Query 209 SICAEFENLWNI 220
IC WNI
Sbjct 286 RICEALATNWNI 297
> pfa:PFL1230w conserved Plasmodium protein
Length=443
Score = 76.3 bits (186), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 43/134 (32%), Positives = 75/134 (55%), Gaps = 2/134 (1%)
Query 43 KLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTY 102
+++Q + ++R DI L++L++S LNK G+L ++I T +G I V P RVP +
Sbjct 263 QIIQSVKNKLENIRLDILFFTLLSLRDSILNKKGKLQIYIYTVNGSFIFVSPLFRVPRNF 322
Query 103 DEFRKLMINLLHTRRIRAVEKNVTLMQVTKNDANLFLPVGSRKIALSTQGRPVDLGDFVQ 162
F+K+M+NLL + +KNV L+++ ND ++ + IA+S +G P D+
Sbjct 323 TLFKKVMLNLLKRGVVYDDQKNV-LLKIIFNDITSYVE-DAVCIAISNKGFPTDVKKLTD 380
Query 163 QFQDTKNPVLFLVG 176
+ + TKN F +
Sbjct 381 KIKQTKNNYFFFIS 394
> cpv:cgd4_2830 Mra1/NEP1 like protein, involved in pre-rRNA processing,
adjacent genes paralogs
Length=216
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 56/197 (28%), Positives = 93/197 (47%), Gaps = 12/197 (6%)
Query 37 EGFQHRKLLQQENRSIA--DVRPDITHQCLIALQESPLNKAGRLC-VFIRTQSGQLIEVH 93
E K+L+++ + I + RPDI H L+ L +SPL K+G L + I G+LI V
Sbjct 20 ELLTEEKVLERDMKLIDSDEFRPDIIHNSLLMLLDSPLCKSGCLSDILILNLDGKLIRVS 79
Query 94 PQLRVPPTYDEFRKLMINLLHTRRIRAV---EKNVTLMQVTKNDANLFLPVGSRKIALST 150
P+ RVP ++ F K+ L + E+N L+ + + +L + LS
Sbjct 80 PKFRVPRSFKVFSKVFSEFLSSPNGELKLPDEENTVLISLLNSSIEEYLSNSEVVVGLSR 139
Query 151 QGRPVDLGDFVQQFQDTK-----NPVLFLVGAVAHANPTTN-SELAEECISIAPCGLSAA 204
+ V L +F + TK + + F++GA A N + IS++ + +
Sbjct 140 MAKKVSLQEFCRNEIATKIRNGADNINFVIGASATNNSCGQFTSKFTHYISLSDVSMPSY 199
Query 205 VCCSSICAEFENLWNIY 221
+CC+ IC+E E L IY
Sbjct 200 ICCTKICSEMEELLGIY 216
> tpv:TP03_0619 hypothetical protein
Length=271
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/142 (30%), Positives = 73/142 (51%), Gaps = 14/142 (9%)
Query 18 RKPRPKTSDYGREGSLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGR 77
+ P PK L LL+ H ++ E+R+ +D RPD+ H CL++LQ+S LNK G+
Sbjct 20 KSPHPK---------LNLLQEHLHFNNIE-EDRNGSDYRPDLVHFCLLSLQDSILNKEGK 69
Query 78 LCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLMINLLHTRR--IRAVEKNVTLMQVTKNDA 135
+ V++ T G + +V RVP + F K+ LH++ +R ++ + V D
Sbjct 70 IQVYLETLQGHVFKVSNSFRVPRAFKVFNKVFSTFLHSKSKDLRTESGDILIESVVNLDQ 129
Query 136 NLFLPVGSRKIALSTQGRPVDL 157
+P S KIA+ + +D+
Sbjct 130 --LIPDSSIKIAIDNRCPVLDI 149
> bbo:BBOV_III002060 17.m07199; hypothetical protein; K14568 essential
for mitotic growth 1
Length=293
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 41/144 (28%), Positives = 70/144 (48%), Gaps = 19/144 (13%)
Query 50 RSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLM 109
R+ +VRPDI H CL+A+ +S LNK + +++ T + Q+ + R+P T+ F K+M
Sbjct 74 RNWKEVRPDILHFCLLAIHDSILNKEKLVQIWVHTLNSQIYRFSSEFRIPRTFKVFNKVM 133
Query 110 INLLHTRRIRAVEKNVTLMQVTKNDANLF-----LPVGSR-----KIALSTQGRPVDLGD 159
LH+ N ++ +D+ +F LP+ + KIA+S +GRP+
Sbjct 134 AKYLHS--------NAKCLKPEGDDSTVFVERMDLPLDNLGNNGIKIAVSNRGRPLIAEK 185
Query 160 FVQQ-FQDTKNPVLFLVGAVAHAN 182
F D +N + V H +
Sbjct 186 FFADILTDKENDIWIYVSLSNHTH 209
> tpv:TP01_0556 hypothetical protein
Length=105
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 47/76 (61%), Gaps = 0/76 (0%)
Query 50 RSIADVRPDITHQCLIALQESPLNKAGRLCVFIRTQSGQLIEVHPQLRVPPTYDEFRKLM 109
R+ + R DI HQ L+ LQ+S LNK L V+IRT +LI+V+P VP T + F L+
Sbjct 13 RNTRESRLDILHQSLLILQDSILNKELCLKVYIRTVDNELIDVNPAFSVPRTLELFEVLI 72
Query 110 INLLHTRRIRAVEKNV 125
+L+ R++++ N+
Sbjct 73 QSLVTNRKVKSTNNNI 88
> dre:449954 im:7141282; si:ch73-37h15.2
Length=549
Score = 31.2 bits (69), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 55/127 (43%), Gaps = 22/127 (17%)
Query 22 PKTSDYGREGSLQLLEGFQHRKLLQQENRSIADVRPDITHQCLIALQESPLNKAGRLCVF 81
P TSD+ R G L ++ G + RK Q ++ + L+ L + L++ + C
Sbjct 133 PNTSDHER-GLLDIMMGTKTRKTKQL-----------LSDEPLVDL--NALDELNQFCPK 178
Query 82 IRTQSGQLIEVHPQLRVP-PTYDEFRKLMINLLHTR-------RIRAVEKNVTLMQVTKN 133
I S +++ +R+ P + EFRK I L H R R+R N + K+
Sbjct 179 ITLASRSARDINKMIRIDLPRFKEFRKQGIELRHGRFSTAENERLRQNVSNFLALTGVKD 238
Query 134 DANLFLP 140
LF P
Sbjct 239 AIKLFHP 245
> tgo:TGME49_116360 hypothetical protein
Length=196
Score = 30.8 bits (68), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 35/84 (41%), Gaps = 17/84 (20%)
Query 27 YGREGSLQLLEGFQHRKLLQQENRSI---ADVRPDITHQCLIALQESPLNKAGRLCVFIR 83
YGR GS EG + + + S+ A P + C++ +L F R
Sbjct 126 YGRRGSGANSEGIALVRGVGPDAESLIVMATYSPPMVSACVVP----------QLTTFFR 175
Query 84 TQSGQLIEVHPQLRVPPTYDEFRK 107
G + P + VPP Y++FRK
Sbjct 176 AYLGLI----PPIAVPPLYEKFRK 195
Lambda K H
0.321 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7266557660
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40