bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3277_orf1
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
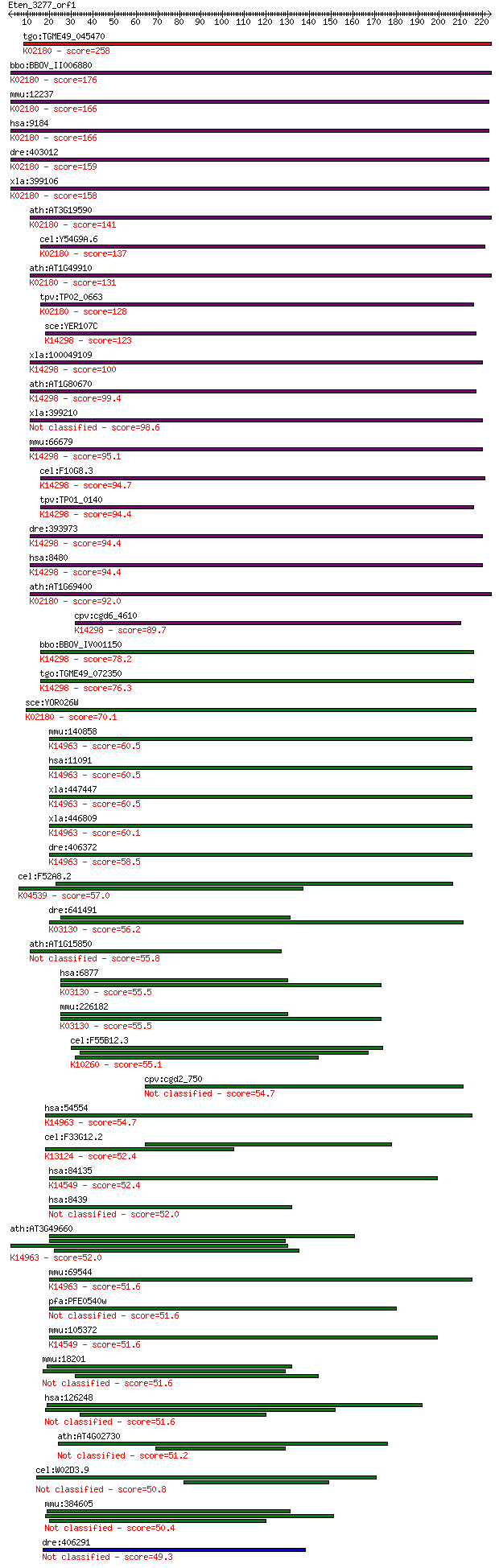
tgo:TGME49_045470 mitotic checkpoint protein BUB3, putative ; ... 258 9e-69
bbo:BBOV_II006880 18.m10036; WD domain/ mitotic checkpoint pro... 176 7e-44
mmu:12237 Bub3, AU019800, AU021329, AU043350, AW146323, C78067... 166 9e-41
hsa:9184 BUB3, BUB3L, hBUB3; budding uninhibited by benzimidaz... 166 9e-41
dre:403012 bub3, MGC101571, zgc:101571; BUB3 budding uninhibit... 159 5e-39
xla:399106 bub3, xbub3; budding uninhibited by benzimidazoles ... 158 1e-38
ath:AT3G19590 WD-40 repeat family protein / mitotic checkpoint... 141 2e-33
cel:Y54G9A.6 bub-3; yeast BUB homolog family member (bub-3); K... 137 4e-32
ath:AT1G49910 WD-40 repeat family protein / mitotic checkpoint... 131 2e-30
tpv:TP02_0663 hypothetical protein; K02180 cell cycle arrest p... 128 1e-29
sce:YER107C GLE2, RAE1; Component of the Nup82 subcomplex of t... 123 5e-28
xla:100049109 rae1, gle2, mig14, mnrp41, mrnp41; RAE1 RNA expo... 100 3e-21
ath:AT1G80670 transducin family protein / WD-40 repeat family ... 99.4 9e-21
xla:399210 rae1/gle2, Rae1; Rae1/Gle2 protein 98.6 2e-20
mmu:66679 Rae1, 3230401I12Rik, 41, D2Ertd342e, MNRP, MNRP41; R... 95.1 2e-19
cel:F10G8.3 npp-17; Nuclear Pore complex Protein family member... 94.7 2e-19
tpv:TP01_0140 mRNA export protein; K14298 mRNA export factor 94.4 3e-19
dre:393973 rae1, MGC56449, zgc:56449, zgc:77723; RAE1 RNA expo... 94.4 3e-19
hsa:8480 RAE1, FLJ30608, MGC117333, MGC126076, MGC126077, MIG1... 94.4 3e-19
ath:AT1G69400 transducin family protein / WD-40 repeat family ... 92.0 2e-18
cpv:cgd6_4610 mRNA export protein ; K14298 mRNA export factor 89.7 8e-18
bbo:BBOV_IV001150 21.m03073; mRNA export protein; K14298 mRNA ... 78.2 2e-14
tgo:TGME49_072350 poly(A)+ RNA export protein, putative ; K142... 76.3 8e-14
sce:YOR026W BUB3; Bub3p; K02180 cell cycle arrest protein BUB3 70.1 6e-12
mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3... 60.5 4e-09
hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPAS... 60.5 4e-09
xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repe... 60.5 5e-09
xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat dom... 60.1 6e-09
dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat d... 58.5 2e-08
cel:F52A8.2 gpb-2; G Protein, Beta subunit family member (gpb-... 57.0 5e-08
dre:641491 taf5, MGC123129, wu:fc38g05, zgc:123129; TAF5 RNA p... 56.2 8e-08
ath:AT1G15850 transducin family protein / WD-40 repeat family ... 55.8 1e-07
hsa:6877 TAF5, TAF2D, TAFII100; TAF5 RNA polymerase II, TATA b... 55.5 1e-07
mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase ... 55.5 2e-07
cel:F55B12.3 sel-10; Suppressor/Enhancer of Lin-12 family memb... 55.1 2e-07
cpv:cgd2_750 bub3'bub3-like protein with WD40 repeats' 54.7 2e-07
hsa:54554 WDR5B, FLJ11287, MGC49879; WD repeat domain 5B; K149... 54.7 2e-07
cel:F33G12.2 hypothetical protein; K13124 mitogen-activated pr... 52.4 1e-06
hsa:84135 UTP15, FLJ12787, NET21; UTP15, U3 small nucleolar ri... 52.4 1e-06
hsa:8439 NSMAF, FAN; neutral sphingomyelinase (N-SMase) activa... 52.0 1e-06
ath:AT3G49660 transducin family protein / WD-40 repeat family ... 52.0 1e-06
mmu:69544 Wdr5b, 2310009C03Rik, AI606931; WD repeat domain 5B;... 51.6 2e-06
pfa:PFE0540w WD-repeat protein, putative 51.6 2e-06
mmu:105372 Utp15, AW544865; UTP15, U3 small nucleolar ribonucl... 51.6 2e-06
mmu:18201 Nsmaf, AA959567, C630007J05, Fan; neutral sphingomye... 51.6 2e-06
hsa:126248 WDR88, PQWD; WD repeat domain 88 51.6
ath:AT4G02730 transducin family protein / WD-40 repeat family ... 51.2 3e-06
cel:W02D3.9 unc-37; UNCoordinated family member (unc-37) 50.8 4e-06
mmu:384605 Wdr88, Gm1429, Pqwd; WD repeat domain 88 50.4
dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin ... 49.3 1e-05
> tgo:TGME49_045470 mitotic checkpoint protein BUB3, putative
; K02180 cell cycle arrest protein BUB3
Length=332
Score = 258 bits (660), Expect = 9e-69, Method: Compositional matrix adjust.
Identities = 121/219 (55%), Positives = 157/219 (71%), Gaps = 3/219 (1%)
Query 8 MLAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCC 67
M +LR EPRD+ISS+C++P+H K++LAA+SWD LR+YDVD ++ + + EF LD C
Sbjct 1 MSIDLRHEPRDSISSLCYAPSHGKSILAATSWDKTLRIYDVDANEQLHKFEFDMPLLDAC 60
Query 68 FLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRA 127
FL D +V GGL V + D+ K+ +G H AV+ R+H PTNL+Y+ GWD V+A
Sbjct 61 FLGDSAKVVIGGLDKQVSLCDLQTEKVVSLGSHTGAVKHCRYHVPTNLVYTAGWDGIVKA 120
Query 128 WDLRSPRE--VGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGAN-LVNAEYRDQI 184
WD R P+ + A LHGK +AMD +D L V DSKKRTY+YD+R G L + ++RDQI
Sbjct 121 WDPRMPQSAPICQAQLHGKAFAMDNSDTYLTVADSKKRTYVYDLRQGPQGLASPDFRDQI 180
Query 185 LKYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQS 223
LKYQIR +RCFPNG GFAA+SIEGRV+WEYFD NPEVQS
Sbjct 181 LKYQIRCLRCFPNGTGFAAASIEGRVAWEYFDMNPEVQS 219
> bbo:BBOV_II006880 18.m10036; WD domain/ mitotic checkpoint protein;
K02180 cell cycle arrest protein BUB3
Length=356
Score = 176 bits (445), Expect = 7e-44, Method: Compositional matrix adjust.
Identities = 94/228 (41%), Positives = 131/228 (57%), Gaps = 8/228 (3%)
Query 2 VEGLNEMLAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDV--DNSDLISQHEF 59
++ N ML + P I+ +CF +N+LA++SWD +++Y++ DN +
Sbjct 18 IDPFNNMLIPIEDPPTGVITRVCF--GKTRNLLASTSWDKTVKLYEIYDDNRGRKLRAYT 75
Query 60 QAAP-LDCCFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYS 118
+P LDC F+ ++A G L V+V DV ++ VG H A VRCV+FH N+I +
Sbjct 76 GGSPALDCSFMEGDKKIAFGNLDNQVNVMDVETGDVTLVGTHGAPVRCVQFHDRLNMIIT 135
Query 119 GGWDNTVRAWDLRSPREVGMA--ALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNG-ANL 175
GGWDN +RA+D R A + GKVY MDL D LVV DS KR YIYD+ G
Sbjct 136 GGWDNKIRAFDPRCDTTSAAADVDIFGKVYCMDLLKDTLVVGDSMKRVYIYDLSRGFIGF 195
Query 176 VNAEYRDQILKYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQS 223
+ +D +LKYQ R ++CFP+ +GFA SIEGRV+WEYF E S
Sbjct 196 STPDTKDGVLKYQYRSIKCFPDNRGFALGSIEGRVAWEYFSKAQEFVS 243
> mmu:12237 Bub3, AU019800, AU021329, AU043350, AW146323, C78067;
budding uninhibited by benzimidazoles 3 homolog (S. cerevisiae);
K02180 cell cycle arrest protein BUB3
Length=326
Score = 166 bits (419), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 88/221 (39%), Positives = 132/221 (59%), Gaps = 6/221 (2%)
Query 2 VEGLNEMLAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQA 61
+ G NE +L P D ISS+ FSPN ++ L SSWD +R+YDV + + +++
Sbjct 1 MTGSNEF--KLNQPPEDGISSVKFSPNTSQ-FLLVSSWDTSVRLYDVPANSMRLKYQHTG 57
Query 62 APLDCCFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGW 121
A LDC F D +GGL + + D++ + + VG HDA +RCV + N++ +G W
Sbjct 58 AVLDCAFY-DPTHAWSGGLDHQLKMHDLNTDQENLVGTHDAPIRCVEYCPEVNVMVTGSW 116
Query 122 DNTVRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR 181
D TV+ WD R+P G + KVY + ++ D+L+V + +R ++D+RN + + R
Sbjct 117 DQTVKLWDPRTPCNAGTFSQPEKVYTLSVSGDRLIVGTAGRRVLVWDLRNMGYV--QQRR 174
Query 182 DQILKYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQ 222
+ LKYQ R +R FPN QG+ SSIEGRV+ EY DP+PEVQ
Sbjct 175 ESSLKYQTRCIRAFPNKQGYVLSSIEGRVAVEYLDPSPEVQ 215
> hsa:9184 BUB3, BUB3L, hBUB3; budding uninhibited by benzimidazoles
3 homolog (yeast); K02180 cell cycle arrest protein BUB3
Length=326
Score = 166 bits (419), Expect = 9e-41, Method: Compositional matrix adjust.
Identities = 88/221 (39%), Positives = 132/221 (59%), Gaps = 6/221 (2%)
Query 2 VEGLNEMLAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQA 61
+ G NE +L P D ISS+ FSPN ++ L SSWD +R+YDV + + +++
Sbjct 1 MTGSNEF--KLNQPPEDGISSVKFSPNTSQ-FLLVSSWDTSVRLYDVPANSMRLKYQHTG 57
Query 62 APLDCCFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGW 121
A LDC F D +GGL + + D++ + + VG HDA +RCV + N++ +G W
Sbjct 58 AVLDCAFY-DPTHAWSGGLDHQLKMHDLNTDQENLVGTHDAPIRCVEYCPEVNVMVTGSW 116
Query 122 DNTVRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR 181
D TV+ WD R+P G + KVY + ++ D+L+V + +R ++D+RN + + R
Sbjct 117 DQTVKLWDPRTPCNAGTFSQPEKVYTLSVSGDRLIVGTAGRRVLVWDLRNMGYV--QQRR 174
Query 182 DQILKYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQ 222
+ LKYQ R +R FPN QG+ SSIEGRV+ EY DP+PEVQ
Sbjct 175 ESSLKYQTRCIRAFPNKQGYVLSSIEGRVAVEYLDPSPEVQ 215
> dre:403012 bub3, MGC101571, zgc:101571; BUB3 budding uninhibited
by benzimidazoles 3 homolog (yeast); K02180 cell cycle
arrest protein BUB3
Length=326
Score = 159 bits (403), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 85/221 (38%), Positives = 130/221 (58%), Gaps = 6/221 (2%)
Query 2 VEGLNEMLAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQA 61
+ G NE +L P D++S++ FSP+ ++ +L SSWD +R+YD + + +++ A
Sbjct 1 MTGANEF--KLAQGPEDSVSAVKFSPSSSQFLLV-SSWDGSVRLYDASANSMRMKYQHLA 57
Query 62 APLDCCFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGW 121
LDC F SD +GGL + D++ + + VG HDA +RCV F N++ +G W
Sbjct 58 PVLDCAF-SDPTHAWSGGLDSQLKTHDLNTDQDTIVGTHDAPIRCVEFCPEVNVLVTGSW 116
Query 122 DNTVRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR 181
D +VR WD R+P G KVY + + D+L+V + +R ++D+RN + + R
Sbjct 117 DQSVRLWDPRTPCNAGTFTQPEKVYTLSVAGDRLIVGTAGRRVLVWDLRNMGYV--QQRR 174
Query 182 DQILKYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQ 222
+ LKYQ R +R FPN QG+ SSIEGRV+ EY DP+ EVQ
Sbjct 175 ESSLKYQTRCIRAFPNKQGYVLSSIEGRVAVEYLDPSLEVQ 215
> xla:399106 bub3, xbub3; budding uninhibited by benzimidazoles
3 homolog; K02180 cell cycle arrest protein BUB3
Length=330
Score = 158 bits (400), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 86/221 (38%), Positives = 130/221 (58%), Gaps = 6/221 (2%)
Query 2 VEGLNEMLAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQA 61
+ G NE +L P D IS++ FSPN ++ L SSWD+ +R+YDV + + +++
Sbjct 7 MTGSNEF--KLNQAPEDGISAVKFSPNTSQ-FLLVSSWDSSVRLYDVPANTMRLKYQHAG 63
Query 62 APLDCCFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGW 121
LDC F D +GGL + + D++ + VG HDA +RCV + N+I +G W
Sbjct 64 PVLDCAFY-DPTHAWSGGLDHQLKMHDLNTDGDTVVGSHDAPIRCVEYCPEVNVIVTGSW 122
Query 122 DNTVRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR 181
D TV+ WD R+P G + KVY + ++ D+L+V + +R ++D+RN + + R
Sbjct 123 DQTVKLWDPRTPCNAGTFSQPDKVYTLSVSGDRLIVGTAGRRVLVWDLRNMGYV--QQRR 180
Query 182 DQILKYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQ 222
+ LKYQ R +R FPN QG+ SSIEGRV+ EY DP+ EVQ
Sbjct 181 ESSLKYQTRCIRAFPNKQGYVLSSIEGRVAVEYLDPSLEVQ 221
> ath:AT3G19590 WD-40 repeat family protein / mitotic checkpoint
protein, putative; K02180 cell cycle arrest protein BUB3
Length=340
Score = 141 bits (356), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 86/219 (39%), Positives = 118/219 (53%), Gaps = 12/219 (5%)
Query 11 ELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLS 70
EL P D IS++ FS N + L SSWD +R+YDV + L + A LDCCF
Sbjct 11 ELSNPPSDGISNLRFSNN--SDHLLVSSWDKRVRLYDVSTNSLKGEFLHGGAVLDCCFHD 68
Query 71 DFNRVAAGG-LTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWD 129
DF+ + G VF+V I +G HD AVRCV + + +G WD TV+ WD
Sbjct 69 DFSGFSVGADYKVRRIVFNVGKEDI--LGTHDKAVRCVEYSYAAGQVITGSWDKTVKCWD 126
Query 130 LRSPR-----EVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQI 184
R +VG +VY+M L +LVV + + IYD+RN + + R+
Sbjct 127 PRGASGPERTQVGTYLQPERVYSMSLVGHRLVVATAGRHVNIYDLRNMSQ--PEQRRESS 184
Query 185 LKYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQS 223
LKYQ R VRC+PNG G+A SS+EGRV+ E+FD + Q+
Sbjct 185 LKYQTRCVRCYPNGTGYALSSVEGRVAMEFFDLSEAAQA 223
> cel:Y54G9A.6 bub-3; yeast BUB homolog family member (bub-3);
K02180 cell cycle arrest protein BUB3
Length=343
Score = 137 bits (344), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 78/209 (37%), Positives = 114/209 (54%), Gaps = 6/209 (2%)
Query 16 PRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEF--QAAPLDCCFLSDFN 73
P IS + F +LAAS WD RVY+V IS+ PL C + +N
Sbjct 22 PFVQISKVQFQREAGSRLLAASGWDGTCRVYEVGKLGDISEKLVFTHGKPLLTCTFAGYN 81
Query 74 RVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSP 133
+VA GG+ V + D+ +Q+G H AVRC+ F+ ++LI SGGWD++V+ WD RS
Sbjct 82 KVAFGGVDHNVKLADIETGNGTQLGSHALAVRCMEFNPMSSLIVSGGWDSSVKLWDARSY 141
Query 134 REVGMAALH--GKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILKYQIRV 191
+ +++ VYAMD+ ++V ++ ++YD R + + RD LKYQ R
Sbjct 142 GNGAIESVNVSSSVYAMDVLKHTILVGTKDRKIFMYDSRKLREPL--QVRDSPLKYQTRA 199
Query 192 VRCFPNGQGFAASSIEGRVSWEYFDPNPE 220
V+ FP G+ F SSIEGRV+ EY D + E
Sbjct 200 VQFFPTGEAFVVSSIEGRVAVEYVDQSGE 228
> ath:AT1G49910 WD-40 repeat family protein / mitotic checkpoint
protein, putative; K02180 cell cycle arrest protein BUB3
Length=339
Score = 131 bits (329), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 77/218 (35%), Positives = 115/218 (52%), Gaps = 10/218 (4%)
Query 11 ELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLS 70
EL P D IS++ FS N + L SSWD +R+YD + + + + A LDCCF
Sbjct 10 ELSNPPSDGISNLRFSNN--SDHLLVSSWDKSVRLYDANGDLMRGEFKHGGAVLDCCFHD 67
Query 71 DFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDL 130
D + + T V D + K +G H+ VRCV + + +G WD T++ WD
Sbjct 68 DSSGFSVCADTK-VRRIDFNAGKEDVLGTHEKPVRCVEYSYAAGQVITGSWDKTIKCWDP 126
Query 131 RSPR-----EVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQIL 185
R ++G +V ++ L ++LVV + + IYD+RN + + R+ L
Sbjct 127 RGASGTERTQIGTYMQPERVNSLSLVGNRLVVATAGRHVNIYDLRNMSQ--PEQRRESSL 184
Query 186 KYQIRVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQS 223
KYQ R VRC+PNG G+A SS+EGRVS E+FD + Q+
Sbjct 185 KYQTRCVRCYPNGTGYALSSVEGRVSMEFFDLSEAAQA 222
> tpv:TP02_0663 hypothetical protein; K02180 cell cycle arrest
protein BUB3
Length=302
Score = 128 bits (322), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 74/205 (36%), Positives = 103/205 (50%), Gaps = 37/205 (18%)
Query 16 PRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSD---LISQHEFQAAPLDCCFLSDF 72
PRD ++ + F + N+LA SSWD ++ YD D + L+ ++++ LD F +
Sbjct 11 PRDVVTKVLFG--NKTNLLAISSWDQTVKFYDADQPNKNRLLFNLDWESTVLDFVFFEND 68
Query 73 NRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRS 132
++A L V + DV VG H+ VRCVR+H PTN + +GGWD VR +DLRS
Sbjct 69 KKMALADLNKNVSLLDVETKNYFTVGLHNGPVRCVRYHEPTNTLITGGWDKKVRVFDLRS 128
Query 133 P--REVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILKYQIR 190
+ V ++GK Y MDL + LVV DS KR
Sbjct 129 SNLKPVVDVDIYGKTYCMDLARNFLVVGDSMKR--------------------------- 161
Query 191 VVRCFPNGQGFAASSIEGRVSWEYF 215
CFP+ G+ SSIEGRV+WEYF
Sbjct 162 ---CFPDATGYVLSSIEGRVAWEYF 183
> sce:YER107C GLE2, RAE1; Component of the Nup82 subcomplex of
the nuclear pore complex; required for polyadenylated RNA export
but not for protein import; homologous to S. pombe Rae1p;
K14298 mRNA export factor
Length=365
Score = 123 bits (308), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 71/206 (34%), Positives = 112/206 (54%), Gaps = 11/206 (5%)
Query 18 DTISSICFSPNHAKNVLAASSWDNMLRVYDVDNS--DLISQHEFQAAPLDCCFLS-DFNR 74
D+IS I FSP + +ASSWD +R++DV N +QHE ++P+ C S D +
Sbjct 37 DSISDIAFSPQQ-DFMFSASSWDGKVRIWDVQNGVPQGRAQHE-SSSPVLCTRWSNDGTK 94
Query 75 VAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHR--PTNL--IYSGGWDNTVRAWDL 130
VA+GG + ++D+ + Q+G H A ++ +RF + P+N I +G WD T++ WD+
Sbjct 95 VASGGCDNALKLYDIASGQTQQIGMHSAPIKVLRFVQCGPSNTECIVTGSWDKTIKYWDM 154
Query 131 RSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILKYQIR 190
R P+ V + +VY+MD LVV +++ I ++ N + A LK+Q R
Sbjct 155 RQPQPVSTVMMPERVYSMDNKQSLLVVATAERHIAIINLANPTTIFKATTSP--LKWQTR 212
Query 191 VVRCFPNGQGFAASSIEGRVSWEYFD 216
V C+ G+A S+EGR S Y D
Sbjct 213 CVACYNEADGYAIGSVEGRCSIRYID 238
> xla:100049109 rae1, gle2, mig14, mnrp41, mrnp41; RAE1 RNA export
1 homolog; K14298 mRNA export factor
Length=368
Score = 100 bits (250), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 72/222 (32%), Positives = 111/222 (50%), Gaps = 17/222 (7%)
Query 11 ELRPEPRDTISSICFSP-NHAKNVLAASSWDNMLRVYDV-DNSDLISQ-HEFQAAP-LDC 66
E+ P D+IS + FSP N L A SW N +R ++V DN I + + P LD
Sbjct 33 EVASPPDDSISCLSFSPPTLPGNFLIAGSWANDVRCWEVQDNGQTIPKAQQMHTGPVLDV 92
Query 67 CFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNL--IYSGGWDNT 124
C+ D +V ++D++ + QV +H+A ++ V + + N I +GGWD +
Sbjct 93 CWSDDGTKVFTASCDKTAKMWDLNSNQSIQVAQHEAPIKTVHWVKAPNYSCIMTGGWDKS 152
Query 125 VRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR--D 182
++ WD RSP + L + Y D+ VV +++ +Y + N +E+R D
Sbjct 153 LKFWDTRSPNPLLTLQLPERCYCADVVYPMAVVATAERGLIVYQLENQP----SEFRRID 208
Query 183 QILKYQIRVVRCFPNGQ----GFAASSIEGRVSWEYFD-PNP 219
LK+Q R V F + Q GFA SIEGRV+ Y + PNP
Sbjct 209 SPLKHQHRCVAIFKDKQNKPTGFALGSIEGRVAIHYINPPNP 250
> ath:AT1G80670 transducin family protein / WD-40 repeat family
protein; K14298 mRNA export factor
Length=349
Score = 99.4 bits (246), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 63/215 (29%), Positives = 104/215 (48%), Gaps = 15/215 (6%)
Query 11 ELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAA-----PLD 65
E+ P P D+ISS+ FSP ++L A+SWDN +R +++ S +A+ P+
Sbjct 19 EVTPSPADSISSLSFSPR--ADILVATSWDNQVRCWEISRSGASLASAPKASISHDQPVL 76
Query 66 C-CFLSDFNRVAAGGLTGGVHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDN 123
C + D V +GG ++ + G + V H+ + + + NL+ +G WD
Sbjct 77 CSAWKDDGTTVFSGGCDKQAKMWPLLSGGQPVTVAMHEGPIAAMAWIPGMNLLATGSWDK 136
Query 124 TVRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR-- 181
T++ WD R V L K Y + + +VV + + +++++N E++
Sbjct 137 TLKYWDTRQQNPVHTQQLPDKCYTLSVKHPLMVVGTADRNLIVFNLQN----PQTEFKRI 192
Query 182 DQILKYQIRVVRCFPNGQGFAASSIEGRVSWEYFD 216
LKYQ R V FP+ QGF SIEGRV + D
Sbjct 193 QSPLKYQTRCVTAFPDQQGFLVGSIEGRVGVHHLD 227
> xla:399210 rae1/gle2, Rae1; Rae1/Gle2 protein
Length=368
Score = 98.6 bits (244), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 71/222 (31%), Positives = 110/222 (49%), Gaps = 17/222 (7%)
Query 11 ELRPEPRDTISSICFSPNH-AKNVLAASSWDNMLRVYDV-DNSDLISQ-HEFQAAPL-DC 66
E+ P D+IS + FSP N L A SW N +R ++V DN I + + P+ D
Sbjct 33 EVASPPDDSISCLSFSPQTLPGNFLIAGSWANDVRCWEVQDNGQTIPKAQQMHTGPVQDV 92
Query 67 CFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNL--IYSGGWDNT 124
C+ D +V ++D++ + Q+ +HDA ++ V + + N I +G WD T
Sbjct 93 CWSDDGTKVFTASCDKTAKMWDLNSNQSIQIAQHDAPIKTVHWVKAPNYSCIMTGSWDKT 152
Query 125 VRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR--D 182
++ WD RSP + L + Y D+ VV +++ +Y + N +E+R D
Sbjct 153 LKFWDTRSPNPLLSIQLPERGYCADVVYPMAVVATAERGLIVYQLENQP----SEFRRID 208
Query 183 QILKYQIRVVRCFPNGQ----GFAASSIEGRVSWEYFD-PNP 219
LK+Q R V F + Q GFA SIEGRV+ Y + PNP
Sbjct 209 SPLKHQHRCVGIFKDKQNKPTGFALGSIEGRVAIHYINPPNP 250
> mmu:66679 Rae1, 3230401I12Rik, 41, D2Ertd342e, MNRP, MNRP41;
RAE1 RNA export 1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 95.1 bits (235), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 66/222 (29%), Positives = 109/222 (49%), Gaps = 17/222 (7%)
Query 11 ELRPEPRDTISSICFSP-NHAKNVLAASSWDNMLRVYDVDNS--DLISQHEFQAAP-LDC 66
E+ P D+I + FSP N L A SW N +R ++V +S + + P LD
Sbjct 33 EVTSSPDDSIGCLSFSPPTLPGNFLIAGSWANDVRCWEVQDSGQTIPKAQQMHTGPVLDV 92
Query 67 CFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNL--IYSGGWDNT 124
C+ D ++V ++D++ + Q+ +HDA V+ + + + N + +G WD T
Sbjct 93 CWSDDGSKVFTASCDKTAKMWDLNSNQAIQIAQHDAPVKTIHWIKAPNYSCVMTGSWDKT 152
Query 125 VRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR--D 182
++ WD RS + + L + Y D+ VV +++ +Y + N +E+R +
Sbjct 153 LKFWDTRSSNPMMVLQLPERCYCADVIYPMAVVATAERGLIVYQLENQP----SEFRRIE 208
Query 183 QILKYQIRVVRCFPNGQ----GFAASSIEGRVSWEYFD-PNP 219
LK+Q R V F + Q GFA SIEGRV+ Y + PNP
Sbjct 209 SPLKHQHRCVAIFKDKQNKPTGFALGSIEGRVAIHYINPPNP 250
> cel:F10G8.3 npp-17; Nuclear Pore complex Protein family member
(npp-17); K14298 mRNA export factor
Length=373
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 70/220 (31%), Positives = 110/220 (50%), Gaps = 17/220 (7%)
Query 16 PRDTISSICFSPN-HAKNVLAASSWDNMLRVY---DVDNSDLISQHEFQAAPLDCCFLSD 71
P DTI I FSP K +LA SWD +RV+ D + + +Q A LD ++ D
Sbjct 41 PEDTIQVIKFSPTPQDKPMLACGSWDGTIRVWMFNDANTFEGKAQQNIPAPILDIAWIED 100
Query 72 FNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNL--IYSGGWDNTVRAWD 129
+++ ++D+ +++ VG HD V+ + N + +G +D T+R WD
Sbjct 101 SSKIFIACADKEARLWDLASNQVAVVGTHDGPVKTCHWINGNNYQCLMTGSFDKTLRFWD 160
Query 130 LRS-PREVGMAALH--GKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILK 186
+++ P + MA + +VYA D+ VV + K +Y++ NG V + Q LK
Sbjct 161 MKNLPNQTQMAQIQLPERVYAADVLYPMAVVALANKHIKVYNLENGPTEVK-DIESQ-LK 218
Query 187 YQIRVVRCFP-----NGQGFAASSIEGRVSWEYFD-PNPE 220
+QIR + F N GFA SIEGRV+ +Y D NP+
Sbjct 219 FQIRCISIFKDKSNQNPAGFALGSIEGRVAVQYVDVANPK 258
> tpv:TP01_0140 mRNA export protein; K14298 mRNA export factor
Length=359
Score = 94.4 bits (233), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 68/213 (31%), Positives = 101/213 (47%), Gaps = 14/213 (6%)
Query 16 PRDTISSICFSPNHAKNVLAASSWDNMLRVYDVD-------NSDLISQHEFQAAPLDC-C 67
P D+IS + +S +L A SWD LR++ V N+D++ + Q AP+ C
Sbjct 21 PNDSISHLRWSTTTNPLLLTAGSWDKTLRIWKVTTGLGNAVNTDMVYTFK-QDAPVLCSA 79
Query 68 FLSDFNRVAAGGLTGGVHVFDVHGAKISQV--GRHDAAVRCVRFHRPTNLIYSGGWDNTV 125
F +D R+ GG T V +D++ + V RH V V + NL+ S WD V
Sbjct 80 FSTDSMRLFGGGCTNNVLAYDLNNPSSTGVVIARHQKPVSGVHWIPQFNLLLSTSWDGGV 139
Query 126 RAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQIL 185
WD R V L KV+A D+ D+ + V DS ++ ++ + + + D L
Sbjct 140 SLWDGRQENPVWSENLGAKVFASDVKDNLMCVADSNRKLSVWSLEKLQHSNSKITIDSSL 199
Query 186 KYQIRVVRCFPNGQ---GFAASSIEGRVSWEYF 215
K QIR + FP+ + G A SSI GR YF
Sbjct 200 KLQIRALSLFPDTKVRSGVAYSSIGGRCVVNYF 232
> dre:393973 rae1, MGC56449, zgc:56449, zgc:77723; RAE1 RNA export
1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 94.4 bits (233), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 65/222 (29%), Positives = 109/222 (49%), Gaps = 17/222 (7%)
Query 11 ELRPEPRDTISSICFSP-NHAKNVLAASSWDNMLRVYDV-DNSDLISQ-HEFQAAP-LDC 66
E+ P ++IS + FSP N L SW N +R ++V DN + + + P LD
Sbjct 33 EVTSPPDESISCLAFSPPTMPGNFLIGGSWANDVRCWEVQDNGQTVPKAQQMHTGPVLDV 92
Query 67 CFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNL--IYSGGWDNT 124
C+ D ++V ++D++ + Q+ +H+ +R + + + N I +G WD T
Sbjct 93 CWSDDGSKVFTASCDKTAKMWDLNSNQAIQIAQHEGPIRTIHWIKAPNYSCIMTGSWDKT 152
Query 125 VRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR--D 182
++ WD RSP + + + Y D+ VV +++ +Y + N +E+R +
Sbjct 153 LKFWDTRSPNPMMSLQMPERCYCADVVYPMAVVATAERGLIVYQLENQP----SEFRRIE 208
Query 183 QILKYQIRVVRCFPNGQ----GFAASSIEGRVSWEYFD-PNP 219
LK+Q R V F + Q GFA SIEGRV+ Y + PNP
Sbjct 209 SPLKHQHRCVAIFKDKQSKPTGFALGSIEGRVAIHYINPPNP 250
> hsa:8480 RAE1, FLJ30608, MGC117333, MGC126076, MGC126077, MIG14,
MRNP41, Mnrp41, dJ481F12.3, dJ800J21.1; RAE1 RNA export
1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 94.4 bits (233), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 66/222 (29%), Positives = 108/222 (48%), Gaps = 17/222 (7%)
Query 11 ELRPEPRDTISSICFSP-NHAKNVLAASSWDNMLRVYDVDNS--DLISQHEFQAAP-LDC 66
E+ P D+I + FSP N L A SW N +R ++V +S + + P LD
Sbjct 33 EVTSSPDDSIGCLSFSPPTLPGNFLIAGSWANDVRCWEVQDSGQTIPKAQQMHTGPVLDV 92
Query 67 CFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNL--IYSGGWDNT 124
C+ D ++V ++D+ + Q+ +HDA V+ + + + N + +G WD T
Sbjct 93 CWSDDGSKVFTASCDKTAKMWDLSSNQAIQIAQHDAPVKTIHWIKAPNYSCVMTGSWDKT 152
Query 125 VRAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYR--D 182
++ WD RS + + L + Y D+ VV +++ +Y + N +E+R +
Sbjct 153 LKFWDTRSSNPMMVLQLPERCYCADVIYPMAVVATAERGLIVYQLENQP----SEFRRIE 208
Query 183 QILKYQIRVVRCFPNGQ----GFAASSIEGRVSWEYFD-PNP 219
LK+Q R V F + Q GFA SIEGRV+ Y + PNP
Sbjct 209 SPLKHQHRCVAIFKDKQNKPTGFALGSIEGRVAIHYINPPNP 250
> ath:AT1G69400 transducin family protein / WD-40 repeat family
protein; K02180 cell cycle arrest protein BUB3
Length=314
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 64/214 (29%), Positives = 106/214 (49%), Gaps = 9/214 (4%)
Query 11 ELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLS 70
E D +S + FSP N L +SWD+ LR+YDV++S L + QAA LDCCF +
Sbjct 7 EFENPIEDAVSRLRFSPQ--SNNLLVASWDSYLRLYDVESSSLSLELNSQAALLDCCFEN 64
Query 71 DFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDL 130
+ +G G + +D++ + +GRHD + + + S G+D ++ WD
Sbjct 65 ESTSFTSGS-DGFIRRYDLNAGTVDTIGRHDDISTSIVYSYEKGEVISTGFDEKIKFWDT 123
Query 131 RSPREVGMAA-LHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILKYQI 189
R + + G V + ++ + LVVC +IYD+RN + Y Q+ + I
Sbjct 124 RQRESLVFSTDAGGAVGCVTVSGNNLVVC-VDASMHIYDLRNLDEAFQS-YASQV-EVPI 180
Query 190 RVVRCFPNGQGFAASSIEGRVSWEYFDPNPEVQS 223
R + P +G+A S++GRV+ ++ PN S
Sbjct 181 RCITSVPYSRGYAVGSVDGRVAVDF--PNTSCSS 212
> cpv:cgd6_4610 mRNA export protein ; K14298 mRNA export factor
Length=333
Score = 89.7 bits (221), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 60/203 (29%), Positives = 96/203 (47%), Gaps = 26/203 (12%)
Query 32 NVLAASSWDNMLRVYDVDNSD-----------LISQHEF-------------QAAP-LDC 66
++LAASSWD + V++V + LIS F +AP LDC
Sbjct 4 SLLAASSWDKSVTVWEVQHMGGNSVNTRFGKFLISPKVFNDLLIVFIGASFQHSAPVLDC 63
Query 67 CFLSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVR 126
SD + +GG + + D+ + +GRHDA + + + + +G WD T++
Sbjct 64 AISSDSRYLFSGGCDNELKMHDMSSRQSQTIGRHDAPISNIFWCDEQKFVVTGSWDKTIK 123
Query 127 AWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILK 186
W+ +S + ++ +VYAMDL LVV + Y+++++N + Q LK
Sbjct 124 FWNGQSQNPIYSLSIPERVYAMDLKYPALVVAAADNAVYVWNLQNITPTPYKRIQTQ-LK 182
Query 187 YQIRVVRCFPNGQGFAASSIEGR 209
Q R + FP+ GFA SIEGR
Sbjct 183 LQPRSISLFPDRTGFAIGSIEGR 205
> bbo:BBOV_IV001150 21.m03073; mRNA export protein; K14298 mRNA
export factor
Length=359
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 57/212 (26%), Positives = 94/212 (44%), Gaps = 12/212 (5%)
Query 16 PRDTISSICFSPNHAKNVLAASSWDNMLRVYDVD-------NSDLISQHEFQAAPLDCCF 68
P D+IS I +S + +L+A SWD +R++ + SD + + +A L CF
Sbjct 20 PDDSISHIRWSHSSNPLLLSAGSWDKTVRLWRISPNIGNTLTSDCVVLYRQEAPILTSCF 79
Query 69 LSDFNRVAAGGLTGGVHVFDV--HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVR 126
D + AGG + V +D+ A V RHD V + + + N + + WD V
Sbjct 80 SDDNTKFFAGGCSNTVMAYDLASRNATGVLVARHDKPVTSIYWVQKYNALLTASWDGRVC 139
Query 127 AWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILK 186
WD R V + K++A + + +++++ + + N D L+
Sbjct 140 LWDGRQSMPVWFDNVDAKIFAFHFRPNVACAGCHNGKIFVWNLDDIQHAKNRVMFDSTLR 199
Query 187 YQIRVVRCFPN---GQGFAASSIEGRVSWEYF 215
QIR + FPN GF SSI GR ++F
Sbjct 200 CQIRSISLFPNLTDKGGFIYSSIGGRAVVKHF 231
> tgo:TGME49_072350 poly(A)+ RNA export protein, putative ; K14298
mRNA export factor
Length=375
Score = 76.3 bits (186), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 61/218 (27%), Positives = 106/218 (48%), Gaps = 21/218 (9%)
Query 16 PRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQA-------APLDC-C 67
PRDTIS + +S + ++L+ +SWDN +RV+ + ++ SQ + A APL C
Sbjct 36 PRDTISQLGWS--NEGSLLSCTSWDNTVRVWQI-SAGFGSQIQAAAKVCMDAQAPLLCST 92
Query 68 FLSDFNRVAAGGLTGGVHVFDVHGAKISQ--VGRHDAAVRCVRFHRPTNLIYSGGWDNTV 125
F N + G V ++D++ + + V +HD V V ++ N+I + WD V
Sbjct 93 FGPSPNHLFVGCCDKTVKLYDLNASSSTPQVVAQHDQPVCSVAWNPIHNVIVTASWDGYV 152
Query 126 RAWDLRSPREVGMAALHGKVYAMDLNDDKLVVCDSKKRTYIYDIRN----GANLVNAEYR 181
R WD + + V ++ GK++ M ++ LV CD+ + + ++ A +
Sbjct 153 RMWDGKQQQPVWQQSVGGKIFRMGVHSPFLVTCDNFRNVNVSNLNTLFTGNAPQQPTKIV 212
Query 182 DQILKYQIRVVRCFPNGQ----GFAASSIEGRVSWEYF 215
+ K Q R + FP+ + G A S+EGRV +F
Sbjct 213 PPLQKLQSRSMGLFPDKEHELPGVAVGSVEGRVGICHF 250
> sce:YOR026W BUB3; Bub3p; K02180 cell cycle arrest protein BUB3
Length=341
Score = 70.1 bits (170), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 61/228 (26%), Positives = 105/228 (46%), Gaps = 26/228 (11%)
Query 9 LAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVD----NSDLISQHEFQAAPL 64
+ ++ P+D IS I P +K++L +SWD L VY D N DL+ ++ L
Sbjct 3 IVQIEQAPKDYISDIKIIP--SKSLLLITSWDGSLTVYKFDIQAKNVDLLQSLRYKHPLL 60
Query 65 DCCFLSDFN-RVAAGGLTGGVHVFDVHGAKISQ-VGRHDAAVRCVRFHR-PTNLIYSGGW 121
C F+ + + ++ G + G + D+ G+ Q + ++A + R + + + + W
Sbjct 61 CCNFIDNTDLQIYVGTVQGEILKVDLIGSPSFQALTNNEANLGICRICKYGDDKLIAASW 120
Query 122 DNTVRAWDLRSPREVGMAAL------------HGKVYAMDLNDDKLVVCDSKKRTYIYDI 169
D + D PR G + K++ MD N +L+V + + + +
Sbjct 121 DGLIEVID---PRNYGDGVIAVKNLNSNNTKVKNKIFTMDTNSSRLIVGMNNSQVQWFRL 177
Query 170 RNGANLVNAEYRDQILKYQIRVVRCFPNGQ-GFAASSIEGRVSWEYFD 216
+ N + LKYQIR V P Q G+A SSI+GRV+ E+FD
Sbjct 178 PLCED-DNGTIEESGLKYQIRDVALLPKEQEGYACSSIDGRVAVEFFD 224
> mmu:140858 Wdr5, 2410008O07Rik, AA408785, AA960360, Big, Big-3;
WD repeat domain 5; K14963 COMPASS component SWD3
Length=334
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 57/203 (28%), Positives = 96/203 (47%), Gaps = 15/203 (7%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRV---YDVDNSDLISQHEFQAAPLDCCFLSDFNRVA 76
+SS+ FSPN LA+SS D ++++ YD IS H+ + D + SD N +
Sbjct 48 VSSVKFSPNG--EWLASSSADKLIKIWGAYDGKFEKTISGHKLGIS--DVAWSSDSNLLV 103
Query 77 AGGLTGGVHVFDVHGAK-ISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPRE 135
+ + ++DV K + + H V C F+ +NLI SG +D +VR WD+++ +
Sbjct 104 SASDDKTLKIWDVSSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKC 163
Query 136 VGMAALHG-KVYAMDLN-DDKLVVCDS-KKRTYIYDIRNGANLVNAEYRDQILKYQIRVV 192
+ H V A+ N D L+V S I+D +G L D + V
Sbjct 164 LKTLPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNP---PVSFV 220
Query 193 RCFPNGQGFAASSIEGRVS-WEY 214
+ PNG+ A++++ + W+Y
Sbjct 221 KFSPNGKYILAATLDNTLKLWDY 243
> hsa:11091 WDR5, BIG-3, SWD3; WD repeat domain 5; K14963 COMPASS
component SWD3
Length=334
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 57/203 (28%), Positives = 96/203 (47%), Gaps = 15/203 (7%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRV---YDVDNSDLISQHEFQAAPLDCCFLSDFNRVA 76
+SS+ FSPN LA+SS D ++++ YD IS H+ + D + SD N +
Sbjct 48 VSSVKFSPNG--EWLASSSADKLIKIWGAYDGKFEKTISGHKLGIS--DVAWSSDSNLLV 103
Query 77 AGGLTGGVHVFDVHGAK-ISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPRE 135
+ + ++DV K + + H V C F+ +NLI SG +D +VR WD+++ +
Sbjct 104 SASDDKTLKIWDVSSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKC 163
Query 136 VGMAALHG-KVYAMDLN-DDKLVVCDS-KKRTYIYDIRNGANLVNAEYRDQILKYQIRVV 192
+ H V A+ N D L+V S I+D +G L D + V
Sbjct 164 LKTLPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNP---PVSFV 220
Query 193 RCFPNGQGFAASSIEGRVS-WEY 214
+ PNG+ A++++ + W+Y
Sbjct 221 KFSPNGKYILAATLDNTLKLWDY 243
> xla:447447 wdr5-b, MGC81485, big-3, swd3, wdr5, xwdr5; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 60.5 bits (145), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 57/203 (28%), Positives = 96/203 (47%), Gaps = 15/203 (7%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRV---YDVDNSDLISQHEFQAAPLDCCFLSDFNRVA 76
+SS+ FSPN LA+SS D ++++ YD IS H+ + D + SD N +
Sbjct 48 VSSVKFSPNG--EWLASSSADKLIKIWGAYDGKFEKTISGHKLGIS--DVAWSSDSNLLV 103
Query 77 AGGLTGGVHVFDVHGAK-ISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPRE 135
+ + ++DV K + + H V C F+ +NLI SG +D +VR WD+++ +
Sbjct 104 SASDDKTLKIWDVSSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKC 163
Query 136 VGMAALHG-KVYAMDLN-DDKLVVCDS-KKRTYIYDIRNGANLVNAEYRDQILKYQIRVV 192
+ H V A+ N D L+V S I+D +G L D + V
Sbjct 164 LKTLPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNP---PVSFV 220
Query 193 RCFPNGQGFAASSIEGRVS-WEY 214
+ PNG+ A++++ + W+Y
Sbjct 221 KFSPNGKYILAATLDNTLKLWDY 243
> xla:446809 wdr5-a, MGC80538, big-3, swd3, xwdr5; WD repeat domain
5; K14963 COMPASS component SWD3
Length=334
Score = 60.1 bits (144), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 56/203 (27%), Positives = 96/203 (47%), Gaps = 15/203 (7%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRV---YDVDNSDLISQHEFQAAPLDCCFLSDFNRVA 76
+SS+ FSPN LA+SS D ++++ YD IS H+ + D + SD N +
Sbjct 48 VSSVKFSPNG--EWLASSSADKLIKIWGAYDGKFEKTISGHKLGIS--DVAWSSDSNLLV 103
Query 77 AGGLTGGVHVFDVHGAK-ISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPRE 135
+ + ++D+ K + + H V C F+ +NLI SG +D +VR WD+++ +
Sbjct 104 SASDDKTLKIWDISSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKC 163
Query 136 VGMAALHG-KVYAMDLN-DDKLVVCDS-KKRTYIYDIRNGANLVNAEYRDQILKYQIRVV 192
+ H V A+ N D L+V S I+D +G L D + V
Sbjct 164 LKTLPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNP---PVSFV 220
Query 193 RCFPNGQGFAASSIEGRVS-WEY 214
+ PNG+ A++++ + W+Y
Sbjct 221 KFSPNGKYILAATLDNTLKLWDY 243
> dre:406372 wdr5, wu:fk47f04, zgc:56591, zgc:76895; WD repeat
domain 5; K14963 COMPASS component SWD3
Length=334
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 56/203 (27%), Positives = 96/203 (47%), Gaps = 15/203 (7%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRV---YDVDNSDLISQHEFQAAPLDCCFLSDFNRVA 76
+SS+ FSP + LA+SS D ++++ YD IS H+ + D + SD N +
Sbjct 48 VSSVKFSP--SGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGIS--DVAWSSDSNLLV 103
Query 77 AGGLTGGVHVFDVHGAK-ISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPRE 135
+ + ++DV K + + H V C F+ +NLI SG +D +VR WD+++ +
Sbjct 104 SASDDKTLKIWDVSSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVRIWDVKTGKC 163
Query 136 VGMAALHG-KVYAMDLN-DDKLVVCDS-KKRTYIYDIRNGANLVNAEYRDQILKYQIRVV 192
+ H V A+ N D L+V S I+D +G L D + V
Sbjct 164 LKTLPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNP---PVSFV 220
Query 193 RCFPNGQGFAASSIEGRVS-WEY 214
+ PNG+ A++++ + W+Y
Sbjct 221 KFSPNGKYILAATLDNTLKLWDY 243
> cel:F52A8.2 gpb-2; G Protein, Beta subunit family member (gpb-2);
K04539 guanine nucleotide binding protein (G protein),
beta 5
Length=356
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 52/196 (26%), Positives = 83/196 (42%), Gaps = 19/196 (9%)
Query 23 ICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAP---LDCCFLSDFNRVAAGG 79
+C + K + +SS D + V+D ++ +H + C F +A GG
Sbjct 72 LCMDWSLDKRHIVSSSQDGKVIVWDGFTTN--KEHALTMPTTWVMACAFSPSSQMIACGG 129
Query 80 LTGGVHVF-----DVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPR 134
L V D K QV H + + C F R NLI +G D+T WD+ S +
Sbjct 130 LDNKCSVVPLSFEDDIIQKKRQVATHTSYMSCCTFLRSDNLILTGSGDSTCAIWDVESGQ 189
Query 135 EVGMAALH-GKVYAMDL----NDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILKYQI 189
+ H G V+A+D+ + + + K + ++DIR+G + + E + I
Sbjct 190 LIQNFHGHTGDVFAIDVPKCDTGNTFISAGADKHSLVWDIRSGQCVQSFEGHEA----DI 245
Query 190 RVVRCFPNGQGFAASS 205
VR PNG FA S
Sbjct 246 NTVRFHPNGDAFATGS 261
Score = 38.9 bits (89), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 32/135 (23%), Positives = 58/135 (42%), Gaps = 4/135 (2%)
Query 6 NEMLAELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQ---HEFQAA 62
++++ + R T C + + N++ S D+ ++DV++ LI H
Sbjct 143 DDIIQKKRQVATHTSYMSCCTFLRSDNLILTGSGDSTCAIWDVESGQLIQNFHGHTGDVF 202
Query 63 PLDCCFLSDFNRVAAGGLTGGVHVFDVH-GAKISQVGRHDAAVRCVRFHRPTNLIYSGGW 121
+D N + G V+D+ G + H+A + VRFH + +G
Sbjct 203 AIDVPKCDTGNTFISAGADKHSLVWDIRSGQCVQSFEGHEADINTVRFHPNGDAFATGSD 262
Query 122 DNTVRAWDLRSPREV 136
D T R +DLR+ R+V
Sbjct 263 DATCRLFDLRADRQV 277
> dre:641491 taf5, MGC123129, wu:fc38g05, zgc:123129; TAF5 RNA
polymerase II, TATA box binding protein (TBP)-associated factor;
K03130 transcription initiation factor TFIID subunit 5
Length=743
Score = 56.2 bits (134), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 54/108 (50%), Gaps = 4/108 (3%)
Query 25 FSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAGGLTGG- 83
F PN N +A S D +R++DV N + + P+ S + A G T G
Sbjct 578 FHPN--SNYVATGSSDRTVRLWDVLNGNCVRIFTGHKGPIHSLAFSPNGKFLASGSTDGR 635
Query 84 VHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDL 130
V ++D+ HG I+++ H + ++F R +I SG DNTVR WD+
Sbjct 636 VLLWDIGHGLMIAELKGHTGTIYALKFSRDGEIIASGSIDNTVRLWDV 683
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 49/197 (24%), Positives = 88/197 (44%), Gaps = 13/197 (6%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNR-VAAG 78
+ + FSP+ +N L +SS D +R++ + + ++ P+ S F +G
Sbjct 489 VYGVSFSPD--RNYLLSSSEDGTIRLWSLQTFTCLVGYKGHNYPVWDTQFSPFGYYFVSG 546
Query 79 GLTGGVHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVG 137
G ++ H + H A V C RFH +N + +G D TVR WD+ + V
Sbjct 547 GHDRVARLWATDHYQPLRIFAGHLADVTCTRFHPNSNYVATGSSDRTVRLWDVLNGNCVR 606
Query 138 MAALH-GKVYAMDL--NDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQI-LKYQIRVVR 193
+ H G ++++ N L + R ++DI +G L+ AE + Y ++ R
Sbjct 607 IFTGHKGPIHSLAFSPNGKFLASGSTDGRVLLWDIGHG--LMIAELKGHTGTIYALKFSR 664
Query 194 CFPNGQGFAASSIEGRV 210
+G+ A+ SI+ V
Sbjct 665 ---DGEIIASGSIDNTV 678
> ath:AT1G15850 transducin family protein / WD-40 repeat family
protein
Length=140
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/123 (30%), Positives = 63/123 (51%), Gaps = 9/123 (7%)
Query 11 ELRPEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSD--LISQHEFQAA---PLD 65
E+ P D+ISS+ FSP ++L A+SWD +R +++ SD + S+ + + P+
Sbjct 20 EITPPATDSISSLSFSPK--ADILVATSWDCQVRCWEITRSDGSIASEPKVSMSHDQPVL 77
Query 66 C-CFLSDFNRVAAGGLTGGVHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDN 123
C + D V GG ++ + GA+ S V HDA + + NL+ +G WD
Sbjct 78 CSAWKDDGTTVFTGGCDKQAKMWPLLSGAQPSTVAMHDAPFNQIAWIPGMNLLVTGSWDK 137
Query 124 TVR 126
T++
Sbjct 138 TLK 140
> hsa:6877 TAF5, TAF2D, TAFII100; TAF5 RNA polymerase II, TATA
box binding protein (TBP)-associated factor, 100kDa; K03130
transcription initiation factor TFIID subunit 5
Length=800
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 52/107 (48%), Gaps = 4/107 (3%)
Query 25 FSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAGGLTGG- 83
F PN N +A S D +R++DV N + + P+ S R A G T G
Sbjct 635 FHPN--SNYVATGSADRTVRLWDVLNGNCVRIFTGHKGPIHSLTFSPNGRFLATGATDGR 692
Query 84 VHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWD 129
V ++D+ HG + ++ H V +RF R ++ SG DNTVR WD
Sbjct 693 VLLWDIGHGLMVGELKGHTDTVCSLRFSRDGEILASGSMDNTVRLWD 739
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 39/153 (25%), Positives = 68/153 (44%), Gaps = 7/153 (4%)
Query 25 FSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPL-DCCFLSDFNRVAAGGLTGG 83
FSP+ +N L +SS D +R++ + + ++ P+ D F +GG
Sbjct 551 FSPD--RNYLLSSSEDGTVRLWSLQTFTCLVGYKGHNYPVWDTQFSPYGYYFVSGGHDRV 608
Query 84 VHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVGMAALH 142
++ H + H A V C RFH +N + +G D TVR WD+ + V + H
Sbjct 609 ARLWATDHYQPLRIFAGHLADVNCTRFHPNSNYVATGSADRTVRLWDVLNGNCVRIFTGH 668
Query 143 -GKVYAMDL--NDDKLVVCDSKKRTYIYDIRNG 172
G ++++ N L + R ++DI +G
Sbjct 669 KGPIHSLTFSPNGRFLATGATDGRVLLWDIGHG 701
> mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase
II, TATA box binding protein (TBP)-associated factor; K03130
transcription initiation factor TFIID subunit 5
Length=801
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 52/107 (48%), Gaps = 4/107 (3%)
Query 25 FSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAGGLTGG- 83
F PN N +A S D +R++DV N + + P+ S R A G T G
Sbjct 636 FHPN--SNYVATGSADRTVRLWDVLNGNCVRIFTGHKGPIHSLTFSPNGRFLATGATDGR 693
Query 84 VHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWD 129
V ++D+ HG + ++ H V +RF R ++ SG DNTVR WD
Sbjct 694 VLLWDIGHGLMVGELKGHTDTVCSLRFSRDGEILASGSMDNTVRLWD 740
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 39/153 (25%), Positives = 68/153 (44%), Gaps = 7/153 (4%)
Query 25 FSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPL-DCCFLSDFNRVAAGGLTGG 83
FSP+ +N L +SS D +R++ + + ++ P+ D F +GG
Sbjct 552 FSPD--RNYLLSSSEDGTVRLWSLQTFTCLVGYKGHNYPVWDTQFSPYGYYFVSGGHDRV 609
Query 84 VHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVGMAALH 142
++ H + H A V C RFH +N + +G D TVR WD+ + V + H
Sbjct 610 ARLWATDHYQPLRIFAGHLADVNCTRFHPNSNYVATGSADRTVRLWDVLNGNCVRIFTGH 669
Query 143 -GKVYAMDL--NDDKLVVCDSKKRTYIYDIRNG 172
G ++++ N L + R ++DI +G
Sbjct 670 KGPIHSLTFSPNGRFLATGATDGRVLLWDIGHG 702
> cel:F55B12.3 sel-10; Suppressor/Enhancer of Lin-12 family member
(sel-10); K10260 F-box and WD-40 domain protein 7
Length=587
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 43/150 (28%), Positives = 68/150 (45%), Gaps = 8/150 (5%)
Query 30 AKNVLAASSWDNMLRVYDVDNS-DLISQHEFQAAPLDCCFLSDFNRVAAGGLTGGVHVFD 88
A ++L S D LRV+DV++ L + H AA C D V +GG V +++
Sbjct 347 AGSILVTGSRDTTLRVWDVESGRHLATLHGHHAAVR--CVQFDGTTVVSGGYDFTVKIWN 404
Query 89 VH-GAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPR-EVGMAALHGK-- 144
H G I + H+ V + F +++ SG D ++R WD P + +A L G
Sbjct 405 AHTGRCIRTLTGHNNRVYSLLFESERSIVCSGSLDTSIRVWDFTRPEGQECVALLQGHTS 464
Query 145 -VYAMDLNDDKLVVCDSKKRTYIYDIRNGA 173
M L + LV C++ ++DI G
Sbjct 465 LTSGMQLRGNILVSCNADSHVRVWDIHEGT 494
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 34/135 (25%), Positives = 66/135 (48%), Gaps = 5/135 (3%)
Query 34 LAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAGGLTGGVHVFDVH-GA 92
+ + S D ++V+ + L+ + + + C ++ + + G + V+DV G
Sbjct 311 IVSGSTDRTVKVWSTVDGSLLHTLQGHTSTVRCMAMAG-SILVTGSRDTTLRVWDVESGR 369
Query 93 KISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVGMAALH-GKVYAMDLN 151
++ + H AAVRCV+F T + SGG+D TV+ W+ + R + H +VY++
Sbjct 370 HLATLHGHHAAVRCVQFDGTT--VVSGGYDFTVKIWNAHTGRCIRTLTGHNNRVYSLLFE 427
Query 152 DDKLVVCDSKKRTYI 166
++ +VC T I
Sbjct 428 SERSIVCSGSLDTSI 442
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 55/114 (48%), Gaps = 6/114 (5%)
Query 32 NVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNR-VAAGGLTGGVHVFD-V 89
+VL S DN L+V+ +D +++ + +S R + +G V V+ V
Sbjct 267 DVLVTGSDDNTLKVWCIDKGEVMYTLVGHTGGVWTSQISQCGRYIVSGSTDRTVKVWSTV 326
Query 90 HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVGMAALHG 143
G+ + + H + VRC+ +++ +G D T+R WD+ S R +A LHG
Sbjct 327 DGSLLHTLQGHTSTVRCMAM--AGSILVTGSRDTTLRVWDVESGRH--LATLHG 376
> cpv:cgd2_750 bub3'bub3-like protein with WD40 repeats'
Length=422
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 44/158 (27%), Positives = 79/158 (50%), Gaps = 12/158 (7%)
Query 64 LDCCFLSDFNRVAAGGLTGGVHVFDVHGAKIS--QVGRHDAAVRCVRFHRPTNLIYSGGW 121
L C F +D + + GG V V ++ + S ++ H A VRC+ ++++ SG W
Sbjct 107 LACKFKND-STIFFGGFNEAVDVINLQDSLYSPRKLVGHLAPVRCLSLLENSSILASGDW 165
Query 122 DNTVRAWDLRSPR---EVGMAALHGKVYAMDL--NDDKLVVCDSKKRTYIYDIRNGANLV 176
+ V + ++ +L GKV+ MD N++ L+V DS K + +++ ++ +
Sbjct 166 NGEVLLTCVNEGSFGSQISKISLPGKVFCMDYSYNEEWLLVGDSLKNMNLINLKKLSSGI 225
Query 177 NAEYRDQI----LKYQIRVVRCFPNGQGFAASSIEGRV 210
A ++ +KYQ+R + + FA SSIEGRV
Sbjct 226 EATTPTEVVPNFMKYQLRNICANKHKDVFATSSIEGRV 263
> hsa:54554 WDR5B, FLJ11287, MGC49879; WD repeat domain 5B; K14963
COMPASS component SWD3
Length=330
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 53/205 (25%), Positives = 96/205 (46%), Gaps = 15/205 (7%)
Query 18 DTISSICFSPNHAKNVLAASSWDNMLRV---YDVDNSDLISQHEFQAAPLDCCFLSDFNR 74
+ +SS+ FSPN LA+SS D ++ + YD + H + + D + SD +R
Sbjct 42 EAVSSVKFSPN--GEWLASSSADRLIIIWGAYDGKYEKTLYGHNLEIS--DVAWSSDSSR 97
Query 75 VAAGGLTGGVHVFDVHGAK-ISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSP 133
+ + + ++DV K + + H V C F+ P+NLI SG +D TV+ W++++
Sbjct 98 LVSASDDKTLKLWDVRSGKCLKTLKGHSNYVFCCNFNPPSNLIISGSFDETVKIWEVKTG 157
Query 134 REVGMAALHGK-VYAMDLN-DDKLVVCDS-KKRTYIYDIRNGANLVNAEYRDQILKYQIR 190
+ + + H V A+ N L+V S I+D +G L D +
Sbjct 158 KCLKTLSAHSDPVSAVHFNCSGSLIVSGSYDGLCRIWDAASGQCLKTLVDDDNP---PVS 214
Query 191 VVRCFPNGQGFAASSIEGRVS-WEY 214
V+ PNG+ ++++ + W+Y
Sbjct 215 FVKFSPNGKYILTATLDNTLKLWDY 239
> cel:F33G12.2 hypothetical protein; K13124 mitogen-activated
protein kinase organizer 1
Length=305
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 62/118 (52%), Gaps = 4/118 (3%)
Query 64 LDCCFLSDFNRVAAGGLTGGVHVFDVH-GAKISQVGRHDAAVRCVRFHRPTNLIYSGGWD 122
LD SD +++AAGG VFDV G ++ + H A V V F+ +++++SG D
Sbjct 62 LDAASSSDNSQIAAGGADRACTVFDVETGKQLRRWRTHGAQVNAVAFNEESSVVFSGSMD 121
Query 123 NTVRAWDLRSPREVGMAALHGK---VYAMDLNDDKLVVCDSKKRTYIYDIRNGANLVN 177
T++A+D RS E + + + ++D+N ++V + +Y IR+G V+
Sbjct 122 CTMQAFDCRSRSEKPIQIFNESTDGILSIDVNGHEIVAGSADGNYRVYSIRDGNMTVD 179
Score = 35.0 bits (79), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 24/90 (26%), Positives = 43/90 (47%), Gaps = 5/90 (5%)
Query 18 DTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQ---HEFQAAPLDCCFLSDFNR 74
D+++S+ F+P+ N L A ++R+ D + L++ H+ LDC L
Sbjct 183 DSVNSVSFTPD--SNCLLAGVMGGIVRLIDKSSGKLLASYKGHQNTEYKLDCRVLQSIEH 240
Query 75 VAAGGLTGGVHVFDVHGAKISQVGRHDAAV 104
VA+G G V+V+ + + I H + V
Sbjct 241 VASGSEDGFVYVYSLLDSHIVSKLEHPSKV 270
> hsa:84135 UTP15, FLJ12787, NET21; UTP15, U3 small nucleolar
ribonucleoprotein, homolog (S. cerevisiae); K14549 U3 small
nucleolar RNA-associated protein 15
Length=518
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 48/185 (25%), Positives = 76/185 (41%), Gaps = 13/185 (7%)
Query 20 ISSICFSPNHAKN-VLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAG 78
+S + FSP N + ASS ++ Y + S+ F+ F D + AG
Sbjct 41 VSKVDFSPQPPYNYAVTASSRIHIYGRYSQEPIKTFSR--FKDTAYCATFRQDGRLLVAG 98
Query 79 GLTGGVHVFDVHG-AKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVG 137
GGV +FD+ G A + Q H AV V F + SG D TV+ WD+ + +E+
Sbjct 99 SEDGGVQLFDISGRAPLRQFEGHTKAVHTVDFTADKYHVVSGADDYTVKLWDIPNSKEIL 158
Query 138 MAALHGKVY----AMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILKYQIRVVR 193
H A LN D + ++D R ++++ E+ + V
Sbjct 159 TFKEHSDYVRCGCASKLNPDLFITGSYDHTVKMFDARTSESVLSVEHGQ-----PVESVL 213
Query 194 CFPNG 198
FP+G
Sbjct 214 LFPSG 218
> hsa:8439 NSMAF, FAN; neutral sphingomyelinase (N-SMase) activation
associated factor
Length=948
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 59/113 (52%), Gaps = 5/113 (4%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAGG 79
+ FSP+ +++VL+ + D L V DV LIS + CF+ D N V +G
Sbjct 839 VCDTAFSPD-SRHVLSTGT-DGCLNVIDVQTGMLISS--MTSDEPQRCFVWDGNSVLSGS 894
Query 80 LTGGVHVFDVHGAKISQ-VGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLR 131
+G + V+D+ GAKIS+ + H AV C+ + + I +GG D + W L+
Sbjct 895 QSGELLVWDLLGAKISERIQGHTGAVTCIWMNEQCSSIITGGEDRQIIFWKLQ 947
> ath:AT3G49660 transducin family protein / WD-40 repeat family
protein; K14963 COMPASS component SWD3
Length=317
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/149 (28%), Positives = 75/149 (50%), Gaps = 10/149 (6%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRVYDVDN-SDLISQ--HEF---QAAPLDCCFLSDFN 73
+SS+ FS + +LA++S D +R Y ++ +D I++ EF + D F SD
Sbjct 27 VSSVKFSSD--GRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSSDAR 84
Query 74 RVAAGGLTGGVHVFDVH-GAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRS 132
+ + + ++DV G+ I + H CV F+ +N+I SG +D TVR WD+ +
Sbjct 85 FIVSASDDKTLKLWDVETGSLIKTLIGHTNYAFCVNFNPQSNMIVSGSFDETVRIWDVTT 144
Query 133 PREVGMAALHG-KVYAMDLNDDKLVVCDS 160
+ + + H V A+D N D ++ S
Sbjct 145 GKCLKVLPAHSDPVTAVDFNRDGSLIVSS 173
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 56/114 (49%), Gaps = 7/114 (6%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCF----LSDFNRV 75
+S + FSPN K +L + DN LR++++ ++ + + C +++ R+
Sbjct 201 VSFVRFSPN-GKFILVGT-LDNTLRLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKRI 258
Query 76 AAGGLTGGVHVFDVHGAKISQ-VGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAW 128
+G VH+++++ K+ Q + H V V H NLI SG D TVR W
Sbjct 259 VSGSEDNCVHMWELNSKKLLQKLEGHTETVMNVACHPTENLIASGSLDKTVRIW 312
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 38/133 (28%), Positives = 61/133 (45%), Gaps = 7/133 (5%)
Query 2 VEGLNEMLAELRPE---PRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHE 58
+ +N+ +AE E + IS + FS + A+ +++AS D L+++DV+ LI
Sbjct 53 INTINDPIAEPVQEFTGHENGISDVAFSSD-ARFIVSASD-DKTLKLWDVETGSLIKTLI 110
Query 59 FQAAPLDCC-FLSDFNRVAAGGLTGGVHVFDVHGAKISQV-GRHDAAVRCVRFHRPTNLI 116
C F N + +G V ++DV K +V H V V F+R +LI
Sbjct 111 GHTNYAFCVNFNPQSNMIVSGSFDETVRIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLI 170
Query 117 YSGGWDNTVRAWD 129
S +D R WD
Sbjct 171 VSSSYDGLCRIWD 183
Score = 37.7 bits (86), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 29/120 (24%), Positives = 52/120 (43%), Gaps = 11/120 (9%)
Query 22 SICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNR----VAA 77
+ C + N N++ + S+D +R++DV + + P+ DFNR + +
Sbjct 116 AFCVNFNPQSNMIVSGSFDETVRIWDVTTGKCLKVLPAHSDPVTAV---DFNRDGSLIVS 172
Query 78 GGLTGGVHVFDV---HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPR 134
G ++D H K + + + V VRF I G DNT+R W++ S +
Sbjct 173 SSYDGLCRIWDSGTGHCVK-TLIDDENPPVSFVRFSPNGKFILVGTLDNTLRLWNISSAK 231
> mmu:69544 Wdr5b, 2310009C03Rik, AI606931; WD repeat domain 5B;
K14963 COMPASS component SWD3
Length=328
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 50/203 (24%), Positives = 97/203 (47%), Gaps = 15/203 (7%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRV---YDVDNSDLISQHEFQAAPLDCCFLSDFNRVA 76
ISS+ FSPN LA+S+ D ++ + YD + + H + + D + SD +R+
Sbjct 42 ISSVKFSPN--GEWLASSAADALIIIWGAYDGNCKKTLYGHSLEIS--DVAWSSDSSRLV 97
Query 77 AGGLTGGVHVFDVHGAK-ISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPRE 135
+ + V+D+ K + + H V C F+ P+NLI SG +D +V+ W++++ +
Sbjct 98 SASDDKTLKVWDMRSGKCLKTLKGHSDFVFCCDFNPPSNLIVSGSFDESVKIWEVKTGKC 157
Query 136 VGMAALHG-KVYAMDLN-DDKLVVCDS-KKRTYIYDIRNGANLVNAEYRDQILKYQIRVV 192
+ + H + A++ N + L+V S I+D +G L + V
Sbjct 158 LKTLSAHSDPISAVNFNCNGSLIVSGSYDGLCRIWDAASGQCLRTLADEGNP---PVSFV 214
Query 193 RCFPNGQGFAASSIEGRVS-WEY 214
+ PNG+ ++++ + W+Y
Sbjct 215 KFSPNGKYILTATLDNTLKLWDY 237
> pfa:PFE0540w WD-repeat protein, putative
Length=526
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 41/165 (24%), Positives = 71/165 (43%), Gaps = 7/165 (4%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPL-DCCFLSDFNRVAAG 78
I ICF N+ N+L S D+ ++++D+ + + P+ F + A+
Sbjct 290 IGDICF--NNKGNILCTCSGDSKIKMWDLVKEKCVHTFKNSTGPIWSLSFHHQGDFFASA 347
Query 79 GLTGGVHVFDVHGAKISQVGR-HDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPR-EV 136
+ + +FD++ + Q+ R H +V V FH + S D T+ WD+RS E
Sbjct 348 SMDQTIRIFDINSLRQRQILRGHVDSVNSVNFHPYFRTLVSASVDKTISIWDMRSGLCEN 407
Query 137 GMAALHGKVYAMDLNDDK--LVVCDSKKRTYIYDIRNGANLVNAE 179
H + N D ++ CDS I+DIR +N +
Sbjct 408 TFYGHHFPCNYSNFNKDANWIISCDSGGVVKIWDIRTNKCFINID 452
> mmu:105372 Utp15, AW544865; UTP15, U3 small nucleolar ribonucleoprotein,
homolog (yeast); K14549 U3 small nucleolar RNA-associated
protein 15
Length=528
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 50/185 (27%), Positives = 75/185 (40%), Gaps = 13/185 (7%)
Query 20 ISSICFSPNHAKN-VLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAG 78
+S + FSP N + ASS ++ Y + S+ F+ F D + AG
Sbjct 41 VSKVDFSPQLPYNYAVTASSRIHIYGRYSQEPVKTFSR--FKDTAYCATFRQDGQLLVAG 98
Query 79 GLTGGVHVFDVHG-AKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVG 137
G V +FD++G A + Q H AV V F + SG D TV+ WD+ + +E+
Sbjct 99 SEDGVVQLFDINGRAPLRQFEGHTKAVHTVDFTADNYHVVSGADDYTVKLWDIPNSKEIL 158
Query 138 MAALHGKVY----AMDLNDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQILKYQIRVVR 193
H A LN D V I+D R N++ E+ + V
Sbjct 159 TFKEHSDYVRCGCASKLNPDLFVTGSYDHTVKIFDARTNKNVLCVEHGQ-----PVESVL 213
Query 194 CFPNG 198
FP+G
Sbjct 214 LFPSG 218
> mmu:18201 Nsmaf, AA959567, C630007J05, Fan; neutral sphingomyelinase
(N-SMase) activation associated factor
Length=920
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 59/114 (51%), Gaps = 5/114 (4%)
Query 19 TISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFLSDFNRVAAG 78
T+ FSP+ ++++L+ D L V DV LIS + CF+ D N V +G
Sbjct 810 TVCDAAFSPD-SRHILSTGV-DGCLNVIDVQTGMLISS--MASEEPQRCFVWDGNSVLSG 865
Query 79 GLTGGVHVFDVHGAKISQ-VGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLR 131
+G + V+D+ GAK+S+ + H AV C+ + + I +GG D + W L+
Sbjct 866 SRSGELLVWDLLGAKVSERIQGHTGAVTCMWMNEQCSSIITGGEDRQIMFWKLQ 919
Score = 35.8 bits (81), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 27/114 (23%), Positives = 52/114 (45%), Gaps = 6/114 (5%)
Query 17 RDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDCCFL-SDFNRV 75
++ ++ I S N + + +S D+ L+++ ++ L F L C L V
Sbjct 633 KEAVTGIAVSCNGSS--VFTTSQDSTLKMFSKESKMLQRSISFSNMALSSCLLLPGDTTV 690
Query 76 AAGGLTGGVHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAW 128
+ V+ + + G + + HD AV + +H + +YSG WD+TV+ W
Sbjct 691 ISSSWDNNVYFYSIAFGRRQDTLMGHDDAVSKICWH--NDRLYSGSWDSTVKVW 742
Score = 30.4 bits (67), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 28/125 (22%), Positives = 52/125 (41%), Gaps = 17/125 (13%)
Query 32 NVLAASSWDNMLRVYDVD---NSDLISQHEFQAAPLDCCFLSDFNRVAAGGLTGGVHVF- 87
+ +SSWDN + Y + D + H+ + + C+ +D R+ +G V V+
Sbjct 688 TTVISSSWDNNVYFYSIAFGRRQDTLMGHDDAVSKI--CWHND--RLYSGSWDSTVKVWS 743
Query 88 ----DVHGAKISQVG-----RHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVGM 138
++ G K Q HD +V + + + L+ SG + V WDL + +
Sbjct 744 GVPAEMPGTKRHQFDLLAELEHDVSVNTINLNAVSTLLVSGTKEGMVNIWDLTTATLLHQ 803
Query 139 AALHG 143
+ H
Sbjct 804 TSCHS 808
> hsa:126248 WDR88, PQWD; WD repeat domain 88
Length=472
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 48/189 (25%), Positives = 89/189 (47%), Gaps = 28/189 (14%)
Query 19 TISSICFSPNHAKNVLAASSWDNMLRVYDVDN--SDLISQHEFQAAPLDCCFLSDFNRVA 76
+I+S CF P+ + +A+ S D ++++DV + + L A +CCF + +
Sbjct 232 SITSCCFDPDSQR--VASVSLDRCIKIWDVTSQATLLTITKAHSNAISNCCFTFSGHFLC 289
Query 77 AGGLTGGVHVFDVH-------GAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWD 129
+ +++VH GA ++ + H+ +V F R ++ + SGG+D TV WD
Sbjct 290 TSSWDKNLKIWNVHTGEFRNCGACVTLMQGHEGSVSSCHFARDSSFLISGGFDRTVAIWD 349
Query 130 L-RSPREVGMAALHGKVYAMDL---NDDKLVVCDSKKRTYIYDIRNGANLVNAEYRDQI- 184
+ R++ + + + MD+ N+ K ++ SK RT L N E D+I
Sbjct 350 VAEGYRKLSLKGHND--WVMDVAISNNKKWILSASKDRTM--------RLWNIEEIDEIP 399
Query 185 --LKYQIRV 191
+KY+ V
Sbjct 400 LVIKYKKAV 408
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/144 (24%), Positives = 62/144 (43%), Gaps = 14/144 (9%)
Query 18 DTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSD---------LISQHEFQAAPLDCCF 68
+ IS+ CF+ + + L SSWD L++++V + L+ HE + C F
Sbjct 274 NAISNCCFT--FSGHFLCTSSWDKNLKIWNVHTGEFRNCGACVTLMQGHEGSVS--SCHF 329
Query 69 LSDFNRVAAGGLTGGVHVFDV-HGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRA 127
D + + +GG V ++DV G + + H+ V V I S D T+R
Sbjct 330 ARDSSFLISGGFDRTVAIWDVAEGYRKLSLKGHNDWVMDVAISNNKKWILSASKDRTMRL 389
Query 128 WDLRSPREVGMAALHGKVYAMDLN 151
W++ E+ + + K + L
Sbjct 390 WNIEEIDEIPLVIKYKKAVGLKLK 413
Score = 30.8 bits (68), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 19/88 (21%), Positives = 40/88 (45%), Gaps = 2/88 (2%)
Query 34 LAASSWDNMLRVYDVDNSDLIS--QHEFQAAPLDCCFLSDFNRVAAGGLTGGVHVFDVHG 91
L + S+D ++++D + ++ +H +A ++C D +RV A V +D+
Sbjct 117 LLSGSYDCTVKLWDPVDGSVVRDFEHRPKAPVVECSITGDSSRVIAASYDKTVRAWDLET 176
Query 92 AKISQVGRHDAAVRCVRFHRPTNLIYSG 119
K+ R+D + +F + SG
Sbjct 177 GKLLWKVRYDTFIVSCKFSPDGKYVVSG 204
> ath:AT4G02730 transducin family protein / WD-40 repeat family
protein
Length=333
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/158 (27%), Positives = 80/158 (50%), Gaps = 6/158 (3%)
Query 24 CFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPL-DCCFLSDFNRVAAGGLTG 82
C ++ N+LA++S D + ++ N LI ++E ++ + D + SD + +
Sbjct 48 CVKFSNDGNLLASASVDKTMILWSATNYSLIHRYEGHSSGISDLAWSSDSHYTCSASDDC 107
Query 83 GVHVFDVHGA-KISQVGR-HDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVGMAA 140
+ ++D + +V R H V CV F+ P+NLI SG +D T+R W++++ + V M
Sbjct 108 TLRIWDARSPYECLKVLRGHTNFVFCVNFNPPSNLIVSGSFDETIRIWEVKTGKCVRMIK 167
Query 141 LHG-KVYAMDLN-DDKLVVCDSKKRT-YIYDIRNGANL 175
H + ++ N D L+V S + I+D + G L
Sbjct 168 AHSMPISSVHFNRDGSLIVSASHDGSCKIWDAKEGTCL 205
Score = 30.8 bits (68), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query 69 LSDFNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAW 128
+++ N G +G V ++ + + + H AA+ CV+F NL+ S D T+ W
Sbjct 12 VANANSTGNAGTSGNVPIYKPY-RHLKTLEGHTAAISCVKFSNDGNLLASASVDKTMILW 70
> cel:W02D3.9 unc-37; UNCoordinated family member (unc-37)
Length=612
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 44/163 (26%), Positives = 76/163 (46%), Gaps = 12/163 (7%)
Query 14 PEPRDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEF--QAAPLDCCFLSD 71
P +D S + S + K L + DN +R +D+ +++H+F Q L CC +
Sbjct 455 PGHQDGASCLDLSKDGTK--LWSGGLDNSVRCWDLAQRKEVAKHDFASQVFSLGCCPNDE 512
Query 72 FNRVAAGGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLR 131
+ VA G V V G + Q+ +H++ V ++F S G DN + AW R
Sbjct 513 W--VAVGMENNYVEVLSTTGKEKYQLTQHESCVLSLKFAHSGKFFISTGKDNALNAW--R 568
Query 132 SPREVGMAAL--HGKVYAMDLN-DDKLVVCDS-KKRTYIYDIR 170
+P + L + V + D++ DD L+V S +K+ +Y +
Sbjct 569 TPYGASLFQLKENSSVLSCDISFDDSLIVTGSGEKKATLYAVE 611
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Query 82 GGVHVFDVHG-AKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPREVGMAA 140
G + ++D+H K+ + H C+ + ++SGG DN+VR WDL +EV
Sbjct 438 GNILIYDIHNKVKVGTLPGHQDGASCLDLSKDGTKLWSGGLDNSVRCWDLAQRKEVAKHD 497
Query 141 LHGKVYAM 148
+V+++
Sbjct 498 FASQVFSL 505
> mmu:384605 Wdr88, Gm1429, Pqwd; WD repeat domain 88
Length=614
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 60/122 (49%), Gaps = 13/122 (10%)
Query 19 TISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDL---ISQHEFQAAPLDCCFLSDFNRV 75
+I+S CF P+ K +A+ S D ++++D+ + I + + A DCCF S + +
Sbjct 379 SITSCCFDPDSQK--VASVSMDRCIKIWDITSRTTLFTIPKAHYNAIS-DCCFTSTGHFL 435
Query 76 AAGGLTGGVHVFDVH-------GAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAW 128
+ +++VH GA ++ + H+ V R ++ + SGG+D TV W
Sbjct 436 CTSSWDKNIKIWNVHTGEFRNRGACVTLMKGHEGCVSSCCIARDSSFLISGGFDKTVAIW 495
Query 129 DL 130
D+
Sbjct 496 DV 497
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/144 (22%), Positives = 64/144 (44%), Gaps = 16/144 (11%)
Query 18 DTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSD---------LISQHEFQAAPLDCCF 68
+ IS CF+ + L SSWD +++++V + L+ HE + CC
Sbjct 421 NAISDCCFTS--TGHFLCTSSWDKNIKIWNVHTGEFRNRGACVTLMKGHEGCVSS--CCI 476
Query 69 LSDFNRVAAGGLTGGVHVFDVHGA--KISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVR 126
D + + +GG V ++DV G K++ G H+ V V I S D+++R
Sbjct 477 ARDSSFLISGGFDKTVAIWDVGGGYRKLALKG-HEDWVTDVAISNNKKWILSASKDSSMR 535
Query 127 AWDLRSPREVGMAALHGKVYAMDL 150
W++ ++ + + + K + +
Sbjct 536 LWNIEEIDKIPLVSENKKAMGLKV 559
Score = 37.7 bits (86), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 26/102 (25%), Positives = 46/102 (45%), Gaps = 4/102 (3%)
Query 20 ISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEF--QAAPLDCCFLSDFNRVAA 77
+SS F N + +SS+D ++++D ++ +I E +A L+C +D R+ A
Sbjct 252 VSSCHFCMNDTR--FLSSSYDCSVKLWDAEDGTVIWDFEPRPKAPVLECSITADNRRIVA 309
Query 78 GGLTGGVHVFDVHGAKISQVGRHDAAVRCVRFHRPTNLIYSG 119
V +DV ++ RHD V C +F + S
Sbjct 310 SSYDKTVRAWDVETGQLLWKVRHDTFVVCCKFSPNGKFVLSA 351
> dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin
p80 (WD repeat containing) subunit B 1
Length=694
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 61/123 (49%), Gaps = 4/123 (3%)
Query 17 RDTISSICFSPNHAKNVLAASSWDNMLRVYDVDNSDLISQHEFQAAPLDC-CFLSDFNRV 75
+ +ISS+ F P LA+ S D+ ++++DV + +++ + C F D +
Sbjct 105 KASISSLDFHP--MGEYLASGSVDSNIKLWDVRRKGCVFRYKGHTQAVRCLAFSPDGKWL 162
Query 76 AAGGLTGGVHVFD-VHGAKISQVGRHDAAVRCVRFHRPTNLIYSGGWDNTVRAWDLRSPR 134
A+ V ++D + G I++ H +AV V+FH L+ SG D TV+ WDL
Sbjct 163 ASASDDSTVKLWDLIAGKMITEFTSHTSAVNVVQFHPNEYLLASGSADRTVKLWDLEKFN 222
Query 135 EVG 137
+G
Sbjct 223 MIG 225
Lambda K H
0.321 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7395169300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40