bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3249_orf2
Length=171
Score E
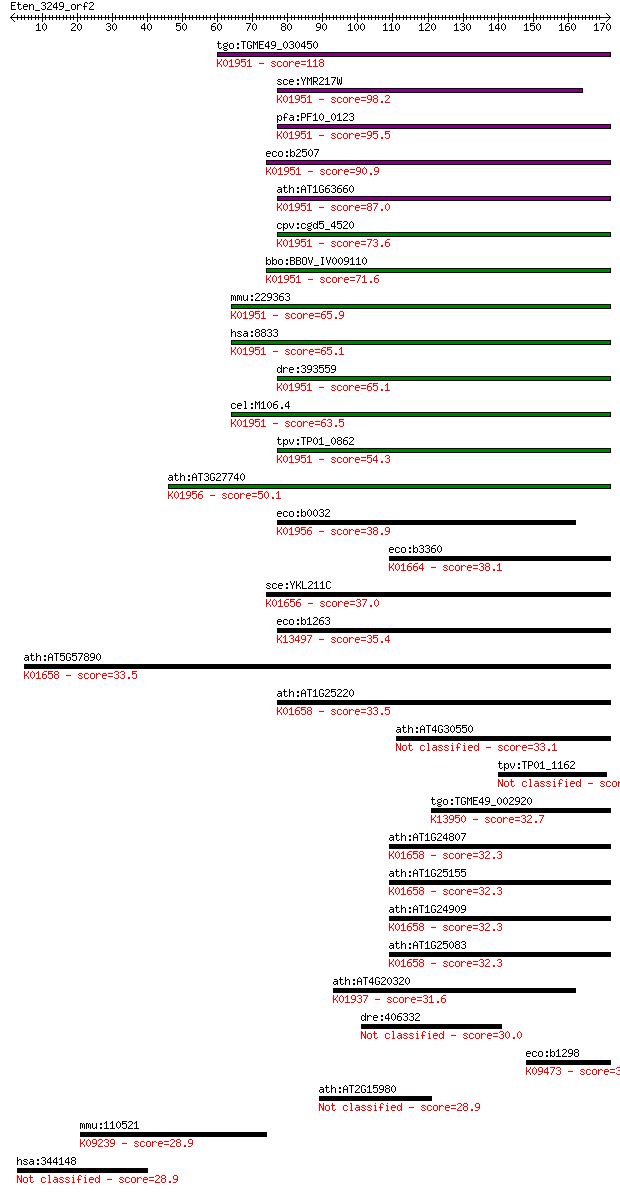
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_030450 GMP synthase, putative (EC:6.3.5.2 6.3.1.5);... 118 1e-26
sce:YMR217W GUA1; GMP synthase, an enzyme that catalyzes the s... 98.2 1e-20
pfa:PF10_0123 GMP synthetase (EC:6.3.5.2); K01951 GMP synthase... 95.5 7e-20
eco:b2507 guaA, ECK2503, JW2491; GMP synthetase (glutamine ami... 90.9 2e-18
ath:AT1G63660 GMP synthase (glutamine-hydrolyzing), putative /... 87.0 3e-17
cpv:cgd5_4520 GMP synthase ; K01951 GMP synthase (glutamine-hy... 73.6 3e-13
bbo:BBOV_IV009110 23.m06540; GMP synthase (EC:6.3.5.2); K01951... 71.6 1e-12
mmu:229363 Gmps, AA591640, AI047208; guanine monophosphate syn... 65.9 7e-11
hsa:8833 GMPS; guanine monphosphate synthetase (EC:6.3.5.2); K... 65.1 1e-10
dre:393559 gmps, MGC66002, sb:cb632, wu:fb76b01, wu:fi05a09, z... 65.1 1e-10
cel:M106.4 hypothetical protein; K01951 GMP synthase (glutamin... 63.5 3e-10
tpv:TP01_0862 GMP synthase; K01951 GMP synthase (glutamine-hyd... 54.3 2e-07
ath:AT3G27740 CARA; CARA (CARBAMOYL PHOSPHATE SYNTHETASE A); c... 50.1 3e-06
eco:b0032 carA, arg, cap, ECK0033, JW0030, pyrA; carbamoyl pho... 38.9 0.009
eco:b3360 pabA, ECK3348, JW3323; aminodeoxychorismate synthase... 38.1 0.014
sce:YKL211C TRP3; Bifunctional enzyme exhibiting both indole-3... 37.0 0.032
eco:b1263 trpD, ECK1257, JW1255, trpGD; fused glutamine amidot... 35.4 0.094
ath:AT5G57890 anthranilate synthase beta subunit, putative (EC... 33.5 0.34
ath:AT1G25220 ASB1; ASB1 (ANTHRANILATE SYNTHASE BETA SUBUNIT 1... 33.5 0.37
ath:AT4G30550 glutamine amidotransferase class-I domain-contai... 33.1 0.52
tpv:TP01_1162 hypothetical protein 33.1 0.52
tgo:TGME49_002920 para-aminobenzoate synthase, putative (EC:4.... 32.7 0.55
ath:AT1G24807 anthranilate synthase beta subunit, putative (EC... 32.3 0.71
ath:AT1G25155 anthranilate synthase beta subunit, putative (EC... 32.3 0.73
ath:AT1G24909 anthranilate synthase beta subunit, putative (EC... 32.3 0.73
ath:AT1G25083 anthranilate synthase beta subunit, putative (EC... 32.3 0.73
ath:AT4G20320 CTP synthase/ catalytic (EC:6.3.4.2); K01937 CTP... 31.6 1.3
dre:406332 crsp7, med26, wu:fe06c07, wu:fi75a09, zgc:55993; co... 30.0 3.7
eco:b1298 puuD, ECK1293, JW1291, ycjL; gamma-Glu-GABA hydrolas... 30.0 4.1
ath:AT2G15980 pentatricopeptide (PPR) repeat-containing protein 28.9 8.1
mmu:110521 Hivep1, Cryabp1; human immunodeficiency virus type ... 28.9 9.2
hsa:344148 NCKAP5, ERIH1, ERIH2, FLJ34870, NAP5; NCK-associate... 28.9 9.9
> tgo:TGME49_030450 GMP synthase, putative (EC:6.3.5.2 6.3.1.5);
K01951 GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2]
Length=569
Score = 118 bits (295), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 59/112 (52%), Positives = 74/112 (66%), Gaps = 7/112 (6%)
Query 60 SRESELSGGRRRPARHMVLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPA 119
S +E GGR VLV DFGSQ S LI+RRLR +GVY+EL C L+E+ P+
Sbjct 13 SAAAEFDGGR-------VLVLDFGSQYSHLIVRRLREIGVYSELRRCDIGLQEIKGFSPS 65
Query 120 AVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
AV+LSGGP+SVYE G+PHL + Q+ ++ +LGICYGMQEI LGG V
Sbjct 66 AVILSGGPASVYEAGSPHLCPSFFAWAQEAKVAVLGICYGMQEICHALGGKV 117
> sce:YMR217W GUA1; GMP synthase, an enzyme that catalyzes the
second step in the biosynthesis of GMP from inosine 5'-phosphate
(IMP); transcription is not subject to regulation by guanine
but is negatively regulated by nutrient starvation (EC:6.3.5.2);
K01951 GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2]
Length=525
Score = 98.2 bits (243), Expect = 1e-20, Method: Composition-based stats.
Identities = 47/87 (54%), Positives = 58/87 (66%), Gaps = 5/87 (5%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
+LV DFGSQ S LI RRLR +YAE+L CT + E+ P V+LSGGP SVY E AP
Sbjct 14 ILVLDFGSQYSHLITRRLREFNIYAEMLPCTQKISELGWT-PKGVILSGGPYSVYAEDAP 72
Query 137 HLRREVWQLLQQQQIPILGICYGMQEI 163
H+ ++ L +PILGICYGMQE+
Sbjct 73 HVDHAIFDL----NVPILGICYGMQEL 95
> pfa:PF10_0123 GMP synthetase (EC:6.3.5.2); K01951 GMP synthase
(glutamine-hydrolysing) [EC:6.3.5.2]
Length=555
Score = 95.5 bits (236), Expect = 7e-20, Method: Composition-based stats.
Identities = 42/95 (44%), Positives = 63/95 (66%), Gaps = 0/95 (0%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
+LV +FGSQ LI++RL + +++E L+++ V+LSGGP SV E G+P
Sbjct 9 ILVLNFGSQYFHLIVKRLNNIKIFSETKDYGVELKDIKDMNIKGVILSGGPYSVTEAGSP 68
Query 137 HLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
HL++EV++ +++IPI GICYGMQEI Q+ G V
Sbjct 69 HLKKEVFEYFLEKKIPIFGICYGMQEIAVQMNGEV 103
> eco:b2507 guaA, ECK2503, JW2491; GMP synthetase (glutamine aminotransferase)
(EC:6.3.5.2 6.3.4.1); K01951 GMP synthase (glutamine-hydrolysing)
[EC:6.3.5.2]
Length=525
Score = 90.9 bits (224), Expect = 2e-18, Method: Composition-based stats.
Identities = 40/98 (40%), Positives = 62/98 (63%), Gaps = 4/98 (4%)
Query 74 RHMVLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEE 133
+H +L+ DFGSQ ++L+ RR+R +GVY EL + T ++ P+ ++LSGGP S EE
Sbjct 7 KHRILILDFGSQYTQLVARRVRELGVYCELWAWDVTEAQIRDFNPSGIILSGGPESTTEE 66
Query 134 GAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+P + V+ + +P+ G+CYGMQ + QLGG V
Sbjct 67 NSPRAPQYVF----EAGVPVFGVCYGMQTMAMQLGGHV 100
> ath:AT1G63660 GMP synthase (glutamine-hydrolyzing), putative
/ glutamine amidotransferase, putative (EC:6.3.5.2); K01951
GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2]
Length=534
Score = 87.0 bits (214), Expect = 3e-17, Method: Composition-based stats.
Identities = 40/95 (42%), Positives = 63/95 (66%), Gaps = 0/95 (0%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
VL+ D+GSQ + LI RR+R++ V++ ++S T++L+ + + P V+LSGGP SV+ AP
Sbjct 11 VLILDYGSQYTHLITRRIRSLNVFSLVISGTSSLKSITSYNPRVVILSGGPHSVHALDAP 70
Query 137 HLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ + + +LGICYG+Q IVQ+LGG V
Sbjct 71 SFPEGFIEWAESNGVSVLGICYGLQLIVQKLGGVV 105
> cpv:cgd5_4520 GMP synthase ; K01951 GMP synthase (glutamine-hydrolysing)
[EC:6.3.5.2]
Length=691
Score = 73.6 bits (179), Expect = 3e-13, Method: Composition-based stats.
Identities = 38/95 (40%), Positives = 54/95 (56%), Gaps = 6/95 (6%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
+++ DFGSQ S LI +R RA+G Y+E+ + L A+ +V SGGPSSVY++ P
Sbjct 13 IIILDFGSQYSHLIAKRFRALGYYSEIALPSTNLNTFNNAK--GIVFSGGPSSVYDDNIP 70
Query 137 HLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
++ L IPILG+CYG + GG V
Sbjct 71 EFNNDILNL----NIPILGLCYGHYIVNIGYGGQV 101
> bbo:BBOV_IV009110 23.m06540; GMP synthase (EC:6.3.5.2); K01951
GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2]
Length=528
Score = 71.6 bits (174), Expect = 1e-12, Method: Composition-based stats.
Identities = 31/98 (31%), Positives = 58/98 (59%), Gaps = 0/98 (0%)
Query 74 RHMVLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEE 133
++++ +FDFGS S ++R LR +G+ EL +E ++ PAAV+LSGG SV +
Sbjct 5 KNIIFIFDFGSHQSGSMVRFLRGLGITCELFPVEKAVETLSGRVPAAVILSGGSESVKDA 64
Query 134 GAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ + +++ ++ + +P+L + Y M + + LGG V
Sbjct 65 DSIKIDKKILEICASKHVPVLALAYAMYALCETLGGKV 102
> mmu:229363 Gmps, AA591640, AI047208; guanine monophosphate synthetase
(EC:6.3.5.2); K01951 GMP synthase (glutamine-hydrolysing)
[EC:6.3.5.2]
Length=693
Score = 65.9 bits (159), Expect = 7e-11, Method: Composition-based stats.
Identities = 38/112 (33%), Positives = 61/112 (54%), Gaps = 8/112 (7%)
Query 64 ELSGGRRRPARH----MVLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPA 119
E +GG + H V++ D G+Q ++I RR+R + V +E+ +
Sbjct 11 ENAGGDLKDGSHHYEGAVVILDAGAQYGKVIDRRVRELFVQSEIFPLETPAFAIKEQGFR 70
Query 120 AVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
A+++SGGP+SVY E AP ++ + + PILGICYGMQ + + GG+V
Sbjct 71 AIIISGGPNSVYAEDAPWFDPAIFTIGK----PILGICYGMQMMNKVFGGTV 118
> hsa:8833 GMPS; guanine monphosphate synthetase (EC:6.3.5.2);
K01951 GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2]
Length=693
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 37/112 (33%), Positives = 61/112 (54%), Gaps = 8/112 (7%)
Query 64 ELSGGRRRPARH----MVLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPA 119
E +GG + H V++ D G+Q ++I RR+R + V +E+ +
Sbjct 11 ENAGGDLKDGHHHYEGAVVILDAGAQYGKVIDRRVRELFVQSEIFPLETPAFAIKEQGFR 70
Query 120 AVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
A+++SGGP+SVY E AP ++ + + P+LGICYGMQ + + GG+V
Sbjct 71 AIIISGGPNSVYAEDAPWFDPAIFTIGK----PVLGICYGMQMMNKVFGGTV 118
> dre:393559 gmps, MGC66002, sb:cb632, wu:fb76b01, wu:fi05a09,
zgc:66002; guanine monphosphate synthetase (EC:6.3.5.2); K01951
GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2]
Length=692
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 34/95 (35%), Positives = 56/95 (58%), Gaps = 4/95 (4%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
V++ D G+Q ++I RR+R + V +E+L + A+++SGGP+SVY E AP
Sbjct 27 VVILDAGAQYGKVIDRRVREMFVQSEILPLETPAFAIREQGFRAIIISGGPNSVYAEDAP 86
Query 137 HLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
++ + + P+LGICYGMQ + + GG+V
Sbjct 87 WFDPAIFTIGK----PVLGICYGMQMMNKVFGGTV 117
> cel:M106.4 hypothetical protein; K01951 GMP synthase (glutamine-hydrolysing)
[EC:6.3.5.2]
Length=792
Score = 63.5 bits (153), Expect = 3e-10, Method: Composition-based stats.
Identities = 40/111 (36%), Positives = 67/111 (60%), Gaps = 15/111 (13%)
Query 64 ELSGGRRRPARHMVLVFDFGSQVSRLIIRRLRAVGVYAEL--LSCTA-TLEEVAAARPAA 120
++S G R + + DFG+Q ++I RR+R + V +E+ L+ TA TL E+ +
Sbjct 101 KVSSGER------IAILDFGAQYGKVIDRRVRELLVQSEMFPLNTTARTLIELGGFK--G 152
Query 121 VVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+++SGGP+SV+E AP + E++ +P+LGICYG Q + + GG+V
Sbjct 153 IIISGGPNSVFEPEAPSIDPEIFTC----GLPVLGICYGFQLMNKLNGGTV 199
> tpv:TP01_0862 GMP synthase; K01951 GMP synthase (glutamine-hydrolysing)
[EC:6.3.5.2]
Length=520
Score = 54.3 bits (129), Expect = 2e-07, Method: Composition-based stats.
Identities = 28/95 (29%), Positives = 47/95 (49%), Gaps = 0/95 (0%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
VLV DFG S ++R +R +GV EL S P+ V L GG SV++E
Sbjct 8 VLVLDFGCTFSSTLVRAVRELGVRCELESLEKFKGLDKKNLPSGVFLVGGNESVFDEDVL 67
Query 137 HLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ + + L+ ++P+L + +GM + + G +
Sbjct 68 SVEKSLLDTLKGHKVPVLSLSFGMMSVAKSFGAKL 102
> ath:AT3G27740 CARA; CARA (CARBAMOYL PHOSPHATE SYNTHETASE A);
carbamoyl-phosphate synthase (glutamine-hydrolyzing)/ carbamoyl-phosphate
synthase/ catalytic; K01956 carbamoyl-phosphate
synthase small subunit [EC:6.3.5.5]
Length=358
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 54/126 (42%), Gaps = 9/126 (7%)
Query 46 CAFLREIVSKMTVYSRESELSGGRRRPARHMVLVFDFGSQVSRLIIRRLRAVGVYAELLS 105
C E V K E + + R + V+ +DFG + + I+RRL + G ++
Sbjct 144 CKSPYEWVDKTNA---EWDFNTNSRDGKSYKVIAYDFG--IKQNILRRLSSYGCQITVVP 198
Query 106 CTATLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQ 165
T E P ++ S GP P+ V +LL + +P+ GIC G Q + Q
Sbjct 199 STFPAAEALKMNPDGILFSNGPGD--PSAVPYAVETVKELLGK--VPVYGICMGHQLLGQ 254
Query 166 QLGGSV 171
LGG
Sbjct 255 ALGGKT 260
> eco:b0032 carA, arg, cap, ECK0033, JW0030, pyrA; carbamoyl phosphate
synthetase small subunit, glutamine amidotransferase
(EC:6.3.5.5); K01956 carbamoyl-phosphate synthase small subunit
[EC:6.3.5.5]
Length=382
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 39/86 (45%), Gaps = 7/86 (8%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
V+ +DFG++ R I+R L G ++ + E+V P + LS GP + AP
Sbjct 194 VVAYDFGAK--RNILRMLVDRGCRLTIVPAQTSAEDVLKMNPDGIFLSNGPG----DPAP 247
Query 137 -HLRREVWQLLQQQQIPILGICYGMQ 161
Q + IP+ GIC G Q
Sbjct 248 CDYAITAIQKFLETDIPVFGICLGHQ 273
> eco:b3360 pabA, ECK3348, JW3323; aminodeoxychorismate synthase,
subunit II (EC:2.6.1.85); K01664 para-aminobenzoate synthetase
component II [EC:2.6.1.85]
Length=187
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 35/63 (55%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
TL ++ A +P +V+S GP + E G + +V + + +PILG+C G Q + Q G
Sbjct 35 TLADIDALKPQKIVISPGPCTPDEAG---ISLDVIRHYAGR-LPILGVCLGHQAMAQAFG 90
Query 169 GSV 171
G V
Sbjct 91 GKV 93
> sce:YKL211C TRP3; Bifunctional enzyme exhibiting both indole-3-glycerol-phosphate
synthase and anthranilate synthase activities,
forms multifunctional hetero-oligomeric anthranilate
synthase:indole-3-glycerol phosphate synthase enzyme complex
with Trp2p (EC:4.1.3.27 4.1.1.48); K01656 anthranilate synthase
/ indole-3-glycerol phosphate synthase [EC:4.1.3.27 4.1.1.48]
Length=484
Score = 37.0 bits (84), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 47/101 (46%), Gaps = 9/101 (8%)
Query 74 RHMVLVFDFGS---QVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSV 130
+H+VL+ ++ S V + + V VY + T+ E+AA P +++S GP
Sbjct 12 KHVVLIDNYDSFTWNVYEYLCQEGAKVSVYR---NDAITVPEIAALNPDTLLISPGPGHP 68
Query 131 YEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ + R+ + + IP+ GIC G Q + GG V
Sbjct 69 KTDSG--ISRDCIRYFTGK-IPVFGICMGQQCMFDVFGGEV 106
> eco:b1263 trpD, ECK1257, JW1255, trpGD; fused glutamine amidotransferase
(component II) of anthranilate synthase/anthranilate
phosphoribosyl transferase (EC:4.1.3.27 2.4.2.18); K13497
anthranilate synthase/phosphoribosyltransferase [EC:4.1.3.27
2.4.2.18]
Length=531
Score = 35.4 bits (80), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 46/100 (46%), Gaps = 10/100 (10%)
Query 77 VLVFDFGSQVSRLIIRRLRAVG----VYAELLSCTATLEEVAAARPAAVVLSGGPSSVYE 132
+L+ D + + +LR+ G +Y + +E +A ++LS GP E
Sbjct 4 ILLLDNIDSFTYNLADQLRSNGHNVVIYRNHIPAQTLIERLATMSNPVLMLSPGPGVPSE 63
Query 133 EGA-PHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
G P L + + ++PI+GIC G Q IV+ GG V
Sbjct 64 AGCMPELLTRL-----RGKLPIIGICLGHQAIVEAYGGYV 98
> ath:AT5G57890 anthranilate synthase beta subunit, putative (EC:4.1.3.27);
K01658 anthranilate synthase component II [EC:4.1.3.27]
Length=273
Score = 33.5 bits (75), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 32/168 (19%), Positives = 76/168 (45%), Gaps = 9/168 (5%)
Query 5 SSLLSICKVKSEYLWLSPAAEAPHIEQEKNPQGSGVIFFPFCAFLREIVSKMTVYSRESE 64
++L + C ++ +Y + + + NP V++ R++++K ++ ES
Sbjct 4 TTLYNSCLLQPKYGFTTRRLNQSLVNSLTNPTRVSVLW----KSRRDVIAKASIEMAESN 59
Query 65 LSGGRRRPARHMVLVFDFGSQVSRLIIRRLRAVGVYAELL-SCTATLEEVAAARPAAVVL 123
+ ++V D + + + + +G + E+ + T+EE+ +P +++
Sbjct 60 SISSVVVNSSGPIIVIDNYDSFTYNLCQYMGELGCHFEVYRNDELTVEELKRKKPRGLLI 119
Query 124 SGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
S GP + + G + V +L +P+ G+C G+Q I + GG +
Sbjct 120 SPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFGGKI 163
> ath:AT1G25220 ASB1; ASB1 (ANTHRANILATE SYNTHASE BETA SUBUNIT
1); anthranilate synthase (EC:4.1.3.27); K01658 anthranilate
synthase component II [EC:4.1.3.27]
Length=276
Score = 33.5 bits (75), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 47/96 (48%), Gaps = 5/96 (5%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELL-SCTATLEEVAAARPAAVVLSGGPSSVYEEGA 135
++V D + + + + +G + E+ + T+EE+ P V++S GP + + G
Sbjct 75 IIVIDNYDSFTYNLCQYMGELGCHFEVYRNDELTVEELKKKNPRGVLISPGPGTPQDSGI 134
Query 136 PHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ V +L +P+ G+C G+Q I + GG +
Sbjct 135 S--LQTVLEL--GPLVPLFGVCMGLQCIGEAFGGKI 166
> ath:AT4G30550 glutamine amidotransferase class-I domain-containing
protein
Length=249
Score = 33.1 bits (74), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 30/62 (48%), Gaps = 1/62 (1%)
Query 111 EEVAAARPAAVVLSGGPSSVYEEGAPHLRR-EVWQLLQQQQIPILGICYGMQEIVQQLGG 169
+E + V+SG P + + ++ EV Q L + +LGIC+G Q I + GG
Sbjct 56 DENDLDKYDGFVISGSPHDAFGDADWIVKLCEVCQKLDHMKKKVLGICFGHQIITRVKGG 115
Query 170 SV 171
+
Sbjct 116 KI 117
> tpv:TP01_1162 hypothetical protein
Length=318
Score = 33.1 bits (74), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 140 REVWQLLQQQQIPILGICYGMQEIVQQLGGS 170
R + +LL + P+ GIC+G Q I Q LG S
Sbjct 90 RHLIRLLFVHKFPMFGICFGFQVISQALGSS 120
> tgo:TGME49_002920 para-aminobenzoate synthase, putative (EC:4.1.3.27
2.6.1.85); K13950 para-aminobenzoate synthetase [EC:2.6.1.85]
Length=988
Score = 32.7 bits (73), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Query 121 VVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+V+S GP +V E H + L++ +PILGIC G Q + GG +
Sbjct 82 IVISPGPGTVENE---HDFGVCSEALREAAVPILGICLGHQGLGHVYGGKI 129
> ath:AT1G24807 anthranilate synthase beta subunit, putative (EC:4.1.3.27);
K01658 anthranilate synthase component II [EC:4.1.3.27]
Length=235
Score = 32.3 bits (72), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 67 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 122
Query 169 GSV 171
G +
Sbjct 123 GKI 125
> ath:AT1G25155 anthranilate synthase beta subunit, putative (EC:4.1.3.27);
K01658 anthranilate synthase component II [EC:4.1.3.27]
Length=222
Score = 32.3 bits (72), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 54 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 109
Query 169 GSV 171
G +
Sbjct 110 GKI 112
> ath:AT1G24909 anthranilate synthase beta subunit, putative (EC:4.1.3.27);
K01658 anthranilate synthase component II [EC:4.1.3.27]
Length=222
Score = 32.3 bits (72), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 54 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 109
Query 169 GSV 171
G +
Sbjct 110 GKI 112
> ath:AT1G25083 anthranilate synthase beta subunit, putative (EC:4.1.3.27);
K01658 anthranilate synthase component II [EC:4.1.3.27]
Length=222
Score = 32.3 bits (72), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 54 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 109
Query 169 GSV 171
G +
Sbjct 110 GKI 112
> ath:AT4G20320 CTP synthase/ catalytic (EC:6.3.4.2); K01937 CTP
synthase [EC:6.3.4.2]
Length=597
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 40/104 (38%), Gaps = 35/104 (33%)
Query 93 RLRAVGVYAELL----SCTATLEEVAAARPAAVVLSGGPSSVYEEGA----PHLRREVWQ 144
R+ VG Y ELL S L + AR +++ +S E+GA P + W+
Sbjct 299 RIAVVGKYTELLDSYLSIHKALLHASVARRKKLIIDWISASDLEQGAKKENPDAYKAAWK 358
Query 145 LLQ---------------------------QQQIPILGICYGMQ 161
LL+ + +IP LGIC GMQ
Sbjct 359 LLKGADGVLVPGGFGSRGVEGKMLAAKYARENRIPYLGICLGMQ 402
> dre:406332 crsp7, med26, wu:fe06c07, wu:fi75a09, zgc:55993;
cofactor required for Sp1 transcriptional activation, subunit
7
Length=589
Score = 30.0 bits (66), Expect = 3.7, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Query 101 AELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAPHLRR 140
A++ S TL+ + + PA V +S GPSSV EG+ HL R
Sbjct 295 AQVPSPLPTLQPLTS--PAQVCISDGPSSVGLEGSLHLHR 332
> eco:b1298 puuD, ECK1293, JW1291, ycjL; gamma-Glu-GABA hydrolase;
K09473 gamma-glutamyl-gamma-aminobutyrate hydrolase [EC:3.5.1.94]
Length=254
Score = 30.0 bits (66), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 148 QQQIPILGICYGMQEIVQQLGGSV 171
+++IPI IC G+QE+V GGS+
Sbjct 105 ERRIPIFAICRGLQELVVATGGSL 128
> ath:AT2G15980 pentatricopeptide (PPR) repeat-containing protein
Length=498
Score = 28.9 bits (63), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 89 LIIRRLRAVGVYAELLSCTATLEEVAAARPAA 120
+++R+LR+ G+ A++ +C A + EV+ R A+
Sbjct 183 MVMRKLRSRGINAQISTCNALITEVSRRRGAS 214
> mmu:110521 Hivep1, Cryabp1; human immunodeficiency virus type
I enhancer binding protein 1; K09239 human immunodeficiency
virus type I enhancer-binding protein
Length=2688
Score = 28.9 bits (63), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 21 SPAAEAPHIEQEKNPQGSGVIFFPFCAFLREIVSKMTVYSRE-----SELSGGRRRPA 73
S AA+ PH+E++K+ QG G +F C R K+ + SEL G + + A
Sbjct 932 SKAAQTPHLEKKKSHQGRGTMF--ECETCRNRYRKLENFENHKKFYCSELHGPKTKAA 987
> hsa:344148 NCKAP5, ERIH1, ERIH2, FLJ34870, NAP5; NCK-associated
protein 5
Length=1909
Score = 28.9 bits (63), Expect = 9.9, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 23/37 (62%), Gaps = 3/37 (8%)
Query 3 QLSSLLSICKVKSEYLWLSPAAEAPHIEQEKNPQGSG 39
+L+S S C ++ + L P+ + P +++E+ PQG G
Sbjct 536 KLTSCASSCPLEMK---LCPSVQTPQVQRERGPQGQG 569
Lambda K H
0.322 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4276754328
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40