bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3220_orf3
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
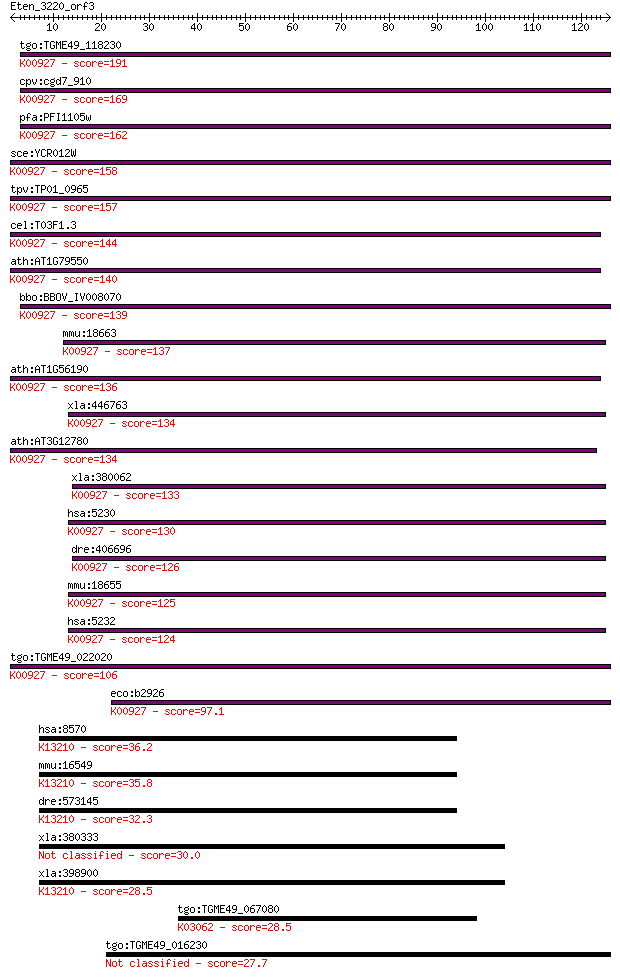
tgo:TGME49_118230 phosphoglycerate kinase, putative (EC:2.7.2.... 191 6e-49
cpv:cgd7_910 phosphoglycerate kinase 1 ; K00927 phosphoglycera... 169 2e-42
pfa:PFI1105w PGK; phosphoglycerate kinase (EC:2.7.2.3); K00927... 162 2e-40
sce:YCR012W PGK1; 3-phosphoglycerate kinase, catalyzes transfe... 158 3e-39
tpv:TP01_0965 phosphoglycerate kinase; K00927 phosphoglycerate... 157 9e-39
cel:T03F1.3 pgk-1; PhosphoGlycerate Kinase family member (pgk-... 144 1e-34
ath:AT1G79550 PGK; PGK (PHOSPHOGLYCERATE KINASE); phosphoglyce... 140 1e-33
bbo:BBOV_IV008070 23.m06017; phosphoglycerate kinase (EC:2.7.2... 139 3e-33
mmu:18663 Pgk2, Pgk-2, Tcp-2; phosphoglycerate kinase 2 (EC:2.... 137 1e-32
ath:AT1G56190 phosphoglycerate kinase, putative; K00927 phosph... 136 1e-32
xla:446763 pgk1, MGC132300, MGC80128, pgk2, pgka; phosphoglyce... 134 6e-32
ath:AT3G12780 PGK1; PGK1 (PHOSPHOGLYCERATE KINASE 1); phosphog... 134 7e-32
xla:380062 pgk1, MGC53000; phosphoglycerate kinase 1 (EC:2.7.2... 133 1e-31
hsa:5230 PGK1, MGC117307, MGC142128, MGC8947, MIG10, PGKA; pho... 130 1e-30
dre:406696 pgk1, wu:fd59b07, wu:fj36g06, zgc:56252, zgc:77899;... 126 2e-29
mmu:18655 Pgk1, MGC118097, Pgk-1; phosphoglycerate kinase 1 (E... 125 4e-29
hsa:5232 PGK2, PGKB, PGKPS, dJ417L20.2; phosphoglycerate kinas... 124 8e-29
tgo:TGME49_022020 phosphoglycerate kinase, putative (EC:2.7.2.... 106 2e-23
eco:b2926 pgk, ECK2922, JW2893; phosphoglycerate kinase (EC:2.... 97.1 1e-20
hsa:8570 KHSRP, FBP2, FUBP2, KSRP, MGC99676; KH-type splicing ... 36.2 0.026
mmu:16549 Khsrp, 6330409F21Rik, Fbp2, Fubp2, Ksrp; KH-type spl... 35.8 0.032
dre:573145 khsrp, MGC163038, fc94c12, wu:fb25b12, wu:fc10e10, ... 32.3 0.35
xla:380333 fubp1, MGC53183; far upstream element (FUSE) bindin... 30.0 1.9
xla:398900 fubp3, MGC68532; far upstream element (FUSE) bindin... 28.5 5.1
tgo:TGME49_067080 26S proteasome subunit 4, putative ; K03062 ... 28.5 6.1
tgo:TGME49_016230 hypothetical protein 27.7 9.7
> tgo:TGME49_118230 phosphoglycerate kinase, putative (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 191 bits (484), Expect = 6e-49, Method: Compositional matrix adjust.
Identities = 94/123 (76%), Positives = 104/123 (84%), Gaps = 0/123 (0%)
Query 3 DANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTA 62
+ANTQICEDK G+PDGWMG+D+GPKTVE A MI RAKT+VWNGPPGVFEM FAKGS A
Sbjct 295 NANTQICEDKTGIPDGWMGVDIGPKTVEKATEMILRAKTLVWNGPPGVFEMSNFAKGSIA 354
Query 63 FAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAAL 122
F AVA AT G +IVGGGDTA+LVE+ G A K+SHVSTGGGASLELLEGK LPGVAAL
Sbjct 355 FCGAVAKATEKGCITIVGGGDTAALVEREGYASKVSHVSTGGGASLELLEGKTLPGVAAL 414
Query 123 SSK 125
S+K
Sbjct 415 SNK 417
> cpv:cgd7_910 phosphoglycerate kinase 1 ; K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=404
Score = 169 bits (427), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 82/123 (66%), Positives = 99/123 (80%), Gaps = 0/123 (0%)
Query 3 DANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTA 62
+AN +I +D G+PDGWMGLD+GP++ K +I +KTIV NGPPGVFEM F+KGS +
Sbjct 281 EANARIIKDSEGIPDGWMGLDIGPESSNSFKELILSSKTIVANGPPGVFEMEKFSKGSRS 340
Query 63 FAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAAL 122
EA+ AAT +G+ +IVGGGDTASLVEKCG A K+SHVSTGGGASLELLEGK LPGV +L
Sbjct 341 MVEALVAATESGSITIVGGGDTASLVEKCGSANKVSHVSTGGGASLELLEGKLLPGVTSL 400
Query 123 SSK 125
SSK
Sbjct 401 SSK 403
> pfa:PFI1105w PGK; phosphoglycerate kinase (EC:2.7.2.3); K00927
phosphoglycerate kinase [EC:2.7.2.3]
Length=416
Score = 162 bits (411), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 77/123 (62%), Positives = 92/123 (74%), Gaps = 0/123 (0%)
Query 3 DANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTA 62
+ANT+ D+ G+PD WMGLD GPK++E K +I +KT++WNGP GVFEMP FAKGS
Sbjct 294 NANTKFVTDEEGIPDNWMGLDAGPKSIENYKDVILTSKTVIWNGPQGVFEMPNFAKGSIE 353
Query 63 FAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAAL 122
V T GA +IVGGGDTASLVE+ ++SHVSTGGGASLELLEGKELPGV AL
Sbjct 354 CLNLVVEVTKKGAITIVGGGDTASLVEQQNKKNEISHVSTGGGASLELLEGKELPGVLAL 413
Query 123 SSK 125
S+K
Sbjct 414 SNK 416
> sce:YCR012W PGK1; 3-phosphoglycerate kinase, catalyzes transfer
of high-energy phosphoryl groups from the acyl phosphate
of 1,3-bisphosphoglycerate to ADP to produce ATP; key enzyme
in glycolysis and gluconeogenesis (EC:2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=416
Score = 158 bits (400), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 77/125 (61%), Positives = 96/125 (76%), Gaps = 0/125 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
+ADANT+ DK G+P GW GLD GP++ +L A +++AKTIVWNGPPGVFE FA G+
Sbjct 291 SADANTKTVTDKEGIPAGWQGLDNGPESRKLFAATVAKAKTIVWNGPPGVFEFEKFAAGT 350
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A + V ++AAG T I+GGGDTA++ +K G+ K+SHVSTGGGASLELLEGKELPGVA
Sbjct 351 KALLDEVVKSSAAGNTVIIGGGDTATVAKKYGVTDKISHVSTGGGASLELLEGKELPGVA 410
Query 121 ALSSK 125
LS K
Sbjct 411 FLSEK 415
> tpv:TP01_0965 phosphoglycerate kinase; K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=415
Score = 157 bits (397), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 77/125 (61%), Positives = 92/125 (73%), Gaps = 0/125 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
+ D ++ + GVPDGWMGLD GPK+VE + AKTIVWNGP GVFE+P F+KGS
Sbjct 291 SNDGPIKLASETQGVPDGWMGLDCGPKSVEKFSEPVLTAKTIVWNGPLGVFELPNFSKGS 350
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
+ V AT GATSI+GGGDTASL E G APK+SHVSTGGGASLELLEGK+LPGV
Sbjct 351 NEVLDLVVKATEKGATSIIGGGDTASLAESTGKAPKLSHVSTGGGASLELLEGKQLPGVV 410
Query 121 ALSSK 125
L+++
Sbjct 411 YLTNR 415
> cel:T03F1.3 pgk-1; PhosphoGlycerate Kinase family member (pgk-1);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 74/123 (60%), Positives = 89/123 (72%), Gaps = 0/123 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DA ++ + GVPDG MGLDVGP++ ++ A I RAKTIVWNGP GVFE FA G+
Sbjct 292 AEDATSKTVTAEEGVPDGHMGLDVGPESSKIFAAAIQRAKTIVWNGPAGVFEFDKFATGT 351
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
+ + V ATAAGA +I+GGGDTA+ +K K+SHVSTGGGASLELLEGK LPGV
Sbjct 352 KSLMDEVVKATAAGAITIIGGGDTATAAKKYNTEDKVSHVSTGGGASLELLEGKVLPGVD 411
Query 121 ALS 123
ALS
Sbjct 412 ALS 414
> ath:AT1G79550 PGK; PGK (PHOSPHOGLYCERATE KINASE); phosphoglycerate
kinase (EC:2.7.7.2 2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=401
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 69/123 (56%), Positives = 88/123 (71%), Gaps = 1/123 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DAN++I A +PDGWMGLD+GP +++ + KTI+WNGP GVFE FA G+
Sbjct 278 APDANSKIVPATA-IPDGWMGLDIGPDSIKTFSEALDTTKTIIWNGPMGVFEFDKFAAGT 336
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A A+ +A + G T+I+GGGD+ + VEK G+A KMSH+STGGGASLELLEGK LPGV
Sbjct 337 EAVAKQLAELSGKGVTTIIGGGDSVAAVEKVGLADKMSHISTGGGASLELLEGKPLPGVL 396
Query 121 ALS 123
AL
Sbjct 397 ALD 399
> bbo:BBOV_IV008070 23.m06017; phosphoglycerate kinase (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=415
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 74/123 (60%), Positives = 85/123 (69%), Gaps = 0/123 (0%)
Query 3 DANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTA 62
DA ++ + G+ D GLD GPKTVEL K +I +TIVWNGP GVFE FA GS A
Sbjct 293 DAPFKVVSEADGISDDSQGLDCGPKTVELLKTVIEGCQTIVWNGPLGVFEFSNFATGSIA 352
Query 63 FAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAAL 122
+ V AT GA S++GGGDTASL E+ G A SHVSTGGGASLEL+EGK LPGV AL
Sbjct 353 ALDIVGEATKRGAISVIGGGDTASLAEQTGRAHLFSHVSTGGGASLELMEGKVLPGVDAL 412
Query 123 SSK 125
SSK
Sbjct 413 SSK 415
> mmu:18663 Pgk2, Pgk-2, Tcp-2; phosphoglycerate kinase 2 (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 66/113 (58%), Positives = 86/113 (76%), Gaps = 0/113 (0%)
Query 12 KAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAAT 71
++G+P GWMGLD GP+++++ ++++AK IVWNGP GVFE AFAKG+ A + V AT
Sbjct 304 ESGIPSGWMGLDCGPESIKINAQIVAQAKLIVWNGPIGVFEWDAFAKGTKALMDEVVKAT 363
Query 72 AAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
+ G +I+GGGDTA+ K G K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 364 SNGCVTIIGGGDTATCCAKWGTEDKVSHVSTGGGASLELLEGKILPGVEALSN 416
> ath:AT1G56190 phosphoglycerate kinase, putative; K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=405
Score = 136 bits (343), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 68/123 (55%), Positives = 87/123 (70%), Gaps = 1/123 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DAN++I A +PDGWMGLD+GP +V+ + +T++WNGP GVFE FAKG+
Sbjct 277 APDANSKIVPASA-IPDGWMGLDIGPDSVKTFNEALDTTQTVIWNGPMGVFEFEKFAKGT 335
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A A +A + G T+I+GGGD+ + VEK G+A MSH+STGGGASLELLEGK LPGV
Sbjct 336 EAVANKLAELSKKGVTTIIGGGDSVAAVEKVGVAGVMSHISTGGGASLELLEGKVLPGVV 395
Query 121 ALS 123
AL
Sbjct 396 ALD 398
> xla:446763 pgk1, MGC132300, MGC80128, pgk2, pgka; phosphoglycerate
kinase 1 (EC:2.7.2.3); K00927 phosphoglycerate kinase
[EC:2.7.2.3]
Length=417
Score = 134 bits (338), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 68/112 (60%), Positives = 80/112 (71%), Gaps = 0/112 (0%)
Query 13 AGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATA 72
AG+PDGWMGLD GP++V+L + RAK IVWNGP GVFE FAKG+ A + V T
Sbjct 305 AGIPDGWMGLDCGPESVKLFVEAVGRAKQIVWNGPVGVFEWDNFAKGTKAVMDKVVEVTG 364
Query 73 AGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +I+GGGDTA+ K K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 365 KGCITIIGGGDTATCCAKWDTEDKVSHVSTGGGASLELLEGKVLPGVDALSN 416
> ath:AT3G12780 PGK1; PGK1 (PHOSPHOGLYCERATE KINASE 1); phosphoglycerate
kinase; K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=481
Score = 134 bits (337), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 66/122 (54%), Positives = 87/122 (71%), Gaps = 1/122 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DAN++I +G+ DGWMGLD+GP +++ + +T++WNGP GVFEM FA G+
Sbjct 353 APDANSKIVP-ASGIEDGWMGLDIGPDSIKTFNEALDTTQTVIWNGPMGVFEMEKFAAGT 411
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A A +A + G T+I+GGGD+ + VEK G+A MSH+STGGGASLELLEGK LPGV
Sbjct 412 EAIANKLAELSEKGVTTIIGGGDSVAAVEKVGVAGVMSHISTGGGASLELLEGKVLPGVI 471
Query 121 AL 122
AL
Sbjct 472 AL 473
> xla:380062 pgk1, MGC53000; phosphoglycerate kinase 1 (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 67/111 (60%), Positives = 79/111 (71%), Gaps = 0/111 (0%)
Query 14 GVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAA 73
G+PDGWMGLD GP++V+L + RAK IVWNGP GVFE FAKG+ A + V T
Sbjct 306 GIPDGWMGLDCGPESVKLFVEAVGRAKQIVWNGPVGVFEWDNFAKGTKAVMDKVVEVTGK 365
Query 74 GATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +I+GGGDTA+ K K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 366 GCITIIGGGDTATCCAKWDTEDKVSHVSTGGGASLELLEGKVLPGVDALSN 416
> hsa:5230 PGK1, MGC117307, MGC142128, MGC8947, MIG10, PGKA; phosphoglycerate
kinase 1 (EC:2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=417
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 65/112 (58%), Positives = 81/112 (72%), Gaps = 0/112 (0%)
Query 13 AGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATA 72
+G+P GWMGLD GP++ + ++RAK IVWNGP GVFE AFA+G+ A + V AT+
Sbjct 305 SGIPAGWMGLDCGPESSKKYAEAVTRAKQIVWNGPVGVFEWEAFARGTKALMDEVVKATS 364
Query 73 AGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +I+GGGDTA+ K K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 365 RGCITIIGGGDTATCCAKWNTEDKVSHVSTGGGASLELLEGKVLPGVDALSN 416
> dre:406696 pgk1, wu:fd59b07, wu:fj36g06, zgc:56252, zgc:77899;
phosphoglycerate kinase 1 (EC:2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=417
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 64/111 (57%), Positives = 77/111 (69%), Gaps = 0/111 (0%)
Query 14 GVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAA 73
G+P GWMGLD GP++ +L ++RAK IVWNGP GVFE FA G+ + V AT
Sbjct 306 GIPAGWMGLDCGPESSKLYAEAVARAKQIVWNGPVGVFEWDNFAHGTKNMMDKVVEATKN 365
Query 74 GATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +I+GGGDTA+ K K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 366 GCITIIGGGDTATCCAKWDTEDKVSHVSTGGGASLELLEGKVLPGVDALSN 416
> mmu:18655 Pgk1, MGC118097, Pgk-1; phosphoglycerate kinase 1
(EC:2.7.2.3); K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 125 bits (313), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 63/112 (56%), Positives = 79/112 (70%), Gaps = 0/112 (0%)
Query 13 AGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATA 72
+G+P GWMGLD G ++ + + RAK IVWNGP GVFE AFA+G+ + + V AT+
Sbjct 305 SGIPAGWMGLDCGTESSKKYAEAVGRAKQIVWNGPVGVFEWEAFARGTKSLMDEVVKATS 364
Query 73 AGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +I+GGGDTA+ K K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 365 RGCITIIGGGDTATCCAKWNTEDKVSHVSTGGGASLELLEGKVLPGVDALSN 416
> hsa:5232 PGK2, PGKB, PGKPS, dJ417L20.2; phosphoglycerate kinase
2 (EC:2.7.2.3); K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 124 bits (311), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 61/112 (54%), Positives = 81/112 (72%), Gaps = 0/112 (0%)
Query 13 AGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATA 72
+G+ GWMGLD GP++ + ++++A+ IVWNGP GVFE AFAKG+ A + + AT+
Sbjct 305 SGISPGWMGLDCGPESNKNHAQVVAQARLIVWNGPLGVFEWDAFAKGTKALMDEIVKATS 364
Query 73 AGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +++GGGDTA+ K K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 365 KGCITVIGGGDTATCCAKWNTEDKVSHVSTGGGASLELLEGKILPGVEALSN 416
> tgo:TGME49_022020 phosphoglycerate kinase, putative (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=551
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 64/125 (51%), Positives = 84/125 (67%), Gaps = 1/125 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
AADA T + +P+GWMGLD GP+T + KT++WNGP G+ E FA G+
Sbjct 424 AADAQTAVVP-VTEIPEGWMGLDNGPQTTARIVEALQDCKTVLWNGPMGMSEYAPFAAGT 482
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A A+A T G T++VGGGD+ + +E+ G++ + SHVSTGGGASLELLEGK LPGVA
Sbjct 483 QEVARALAEITRRGGTTVVGGGDSVAAIERLGLSNEFSHVSTGGGASLELLEGKTLPGVA 542
Query 121 ALSSK 125
ALS +
Sbjct 543 ALSDE 547
> eco:b2926 pgk, ECK2922, JW2893; phosphoglycerate kinase (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=387
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 50/104 (48%), Positives = 68/104 (65%), Gaps = 3/104 (2%)
Query 22 LDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAAGATSIVGG 81
LD+G + + ++ AKTI+WNGP GVFE P F KG+ A A+A + A SI GG
Sbjct 284 LDIGDASAQELAEILKNAKTILWNGPVGVFEFPNFRKGTEIVANAIADSEA---FSIAGG 340
Query 82 GDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSSK 125
GDT + ++ G+A K+S++STGGGA LE +EGK LP VA L +
Sbjct 341 GDTLAAIDLFGIADKISYISTGGGAFLEFVEGKVLPAVAMLEER 384
> hsa:8570 KHSRP, FBP2, FUBP2, KSRP, MGC99676; KH-type splicing
regulatory protein; K13210 far upstream element-binding protein
Length=711
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 38/87 (43%), Gaps = 0/87 (0%)
Query 7 QICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEA 66
QI D G+P+ + L P++V+ AK M+ + GPPG F A + E
Sbjct 179 QISPDSGGLPERSVSLTGAPESVQKAKMMLDDIVSRGRGGPPGQFHDNANGGQNGTVQEI 238
Query 67 VAAATAAGATSIVGGGDTASLVEKCGM 93
+ A AG GG L E+ G+
Sbjct 239 MIPAGKAGLVIGKGGETIKQLQERAGV 265
> mmu:16549 Khsrp, 6330409F21Rik, Fbp2, Fubp2, Ksrp; KH-type splicing
regulatory protein; K13210 far upstream element-binding
protein
Length=748
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 38/87 (43%), Gaps = 0/87 (0%)
Query 7 QICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEA 66
QI D G+P+ + L P++V+ AK M+ + GPPG F A + E
Sbjct 180 QISPDSGGLPERSVSLTGAPESVQKAKMMLDDIVSRGRGGPPGQFHDNANGGQNGTVQEI 239
Query 67 VAAATAAGATSIVGGGDTASLVEKCGM 93
+ A AG GG L E+ G+
Sbjct 240 MIPAGKAGLVIGKGGETIKQLQERAGV 266
> dre:573145 khsrp, MGC163038, fc94c12, wu:fb25b12, wu:fc10e10,
wu:fc94c12, zgc:163038; KH-type splicing regulatory protein;
K13210 far upstream element-binding protein
Length=666
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 36/87 (41%), Gaps = 3/87 (3%)
Query 7 QICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEA 66
QI D G+PD + + GP+ ++ AK M+ + PP E GS E
Sbjct 141 QIAPDSGGLPDRSVSITGGPEAIQKAKMMLDDIVSRGRGTPPSFHES---TNGSGHMQEM 197
Query 67 VAAATAAGATSIVGGGDTASLVEKCGM 93
V A AG GG L E+ G+
Sbjct 198 VIPAGKAGLIIGKGGETIKQLQERAGV 224
> xla:380333 fubp1, MGC53183; far upstream element (FUSE) binding
protein 1
Length=653
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 32/100 (32%), Positives = 43/100 (43%), Gaps = 12/100 (12%)
Query 7 QICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS---TAF 63
QI D AG+PD L P +V+ AK ++ + IV G P P F G A
Sbjct 123 QIAPDSAGMPDRSCMLTGSPDSVQAAKRLLDQ---IVEKGRP----TPGFHHGEGSGNAV 175
Query 64 AEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTG 103
E + A+ AG GG L E+ G+ KM + G
Sbjct 176 QEIMIPASKAGLVIGKGGETIKQLQERAGV--KMVMIQDG 213
> xla:398900 fubp3, MGC68532; far upstream element (FUSE) binding
protein 3; K13210 far upstream element-binding protein
Length=546
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 42/97 (43%), Gaps = 6/97 (6%)
Query 7 QICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEA 66
QI D G+P+ L P+++E AK ++ + NGP +M GS+ E
Sbjct 113 QIAPDSGGMPERPCVLTGTPESIEQAKRLLGQIVDRCRNGPGFHNDM----DGSSTVQEI 168
Query 67 VAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTG 103
+ A+ G GG L E+ G+ KM + G
Sbjct 169 LIPASKVGLVIGKGGETIKQLQERTGV--KMIMIQDG 203
> tgo:TGME49_067080 26S proteasome subunit 4, putative ; K03062
26S proteasome regulatory subunit T2
Length=441
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 13/63 (20%)
Query 36 ISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAAGATSIVGGGDTASLVEK-CGMA 94
IS K ++ GPPG G T A+AVA T+A +VG + L++K G
Sbjct 217 ISPPKGVILYGPPGT--------GKTLLAKAVANETSATFLRVVG----SELIQKYLGDG 264
Query 95 PKM 97
PK+
Sbjct 265 PKL 267
> tgo:TGME49_016230 hypothetical protein
Length=142
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 48/110 (43%), Gaps = 14/110 (12%)
Query 21 GLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAAGATSIVG 80
G D P+ KA+ R +T + VF + STA E ++A A S+
Sbjct 38 GRDARPREKN-RKAVCRRTETEIC-----VFVVGLREPSSTARLEFLSAGLAVPRVSLQR 91
Query 81 GGDTASLVEK-----CGMAPKMSHVSTGGGASLELLEGKELPGVAALSSK 125
TASLV + CG + G G SLE+ G++ PG ++ S+
Sbjct 92 LLATASLVSRPLFPCCGSKETLFD---GFGESLEMRVGRQSPGASSFLSR 138
Lambda K H
0.311 0.128 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40