bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3212_orf1
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
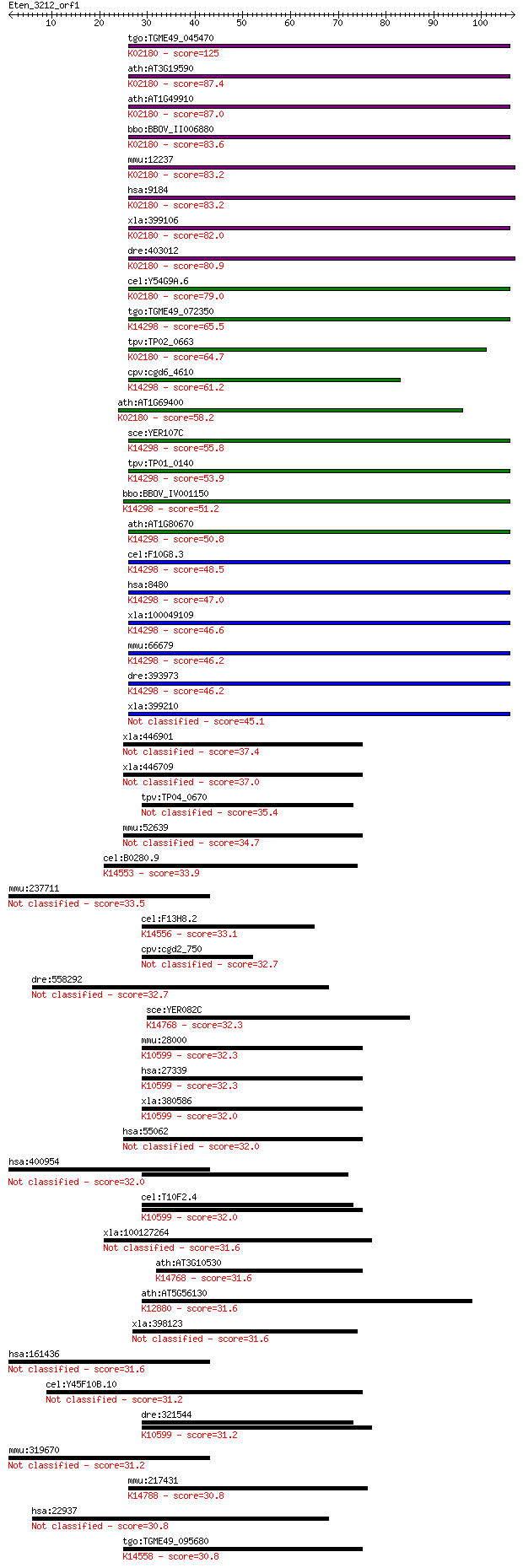
tgo:TGME49_045470 mitotic checkpoint protein BUB3, putative ; ... 125 4e-29
ath:AT3G19590 WD-40 repeat family protein / mitotic checkpoint... 87.4 9e-18
ath:AT1G49910 WD-40 repeat family protein / mitotic checkpoint... 87.0 1e-17
bbo:BBOV_II006880 18.m10036; WD domain/ mitotic checkpoint pro... 83.6 1e-16
mmu:12237 Bub3, AU019800, AU021329, AU043350, AW146323, C78067... 83.2 2e-16
hsa:9184 BUB3, BUB3L, hBUB3; budding uninhibited by benzimidaz... 83.2 2e-16
xla:399106 bub3, xbub3; budding uninhibited by benzimidazoles ... 82.0 4e-16
dre:403012 bub3, MGC101571, zgc:101571; BUB3 budding uninhibit... 80.9 1e-15
cel:Y54G9A.6 bub-3; yeast BUB homolog family member (bub-3); K... 79.0 4e-15
tgo:TGME49_072350 poly(A)+ RNA export protein, putative ; K142... 65.5 3e-11
tpv:TP02_0663 hypothetical protein; K02180 cell cycle arrest p... 64.7 7e-11
cpv:cgd6_4610 mRNA export protein ; K14298 mRNA export factor 61.2 7e-10
ath:AT1G69400 transducin family protein / WD-40 repeat family ... 58.2 6e-09
sce:YER107C GLE2, RAE1; Component of the Nup82 subcomplex of t... 55.8 3e-08
tpv:TP01_0140 mRNA export protein; K14298 mRNA export factor 53.9 1e-07
bbo:BBOV_IV001150 21.m03073; mRNA export protein; K14298 mRNA ... 51.2 9e-07
ath:AT1G80670 transducin family protein / WD-40 repeat family ... 50.8 1e-06
cel:F10G8.3 npp-17; Nuclear Pore complex Protein family member... 48.5 5e-06
hsa:8480 RAE1, FLJ30608, MGC117333, MGC126076, MGC126077, MIG1... 47.0 1e-05
xla:100049109 rae1, gle2, mig14, mnrp41, mrnp41; RAE1 RNA expo... 46.6 2e-05
mmu:66679 Rae1, 3230401I12Rik, 41, D2Ertd342e, MNRP, MNRP41; R... 46.2 2e-05
dre:393973 rae1, MGC56449, zgc:56449, zgc:77723; RAE1 RNA expo... 46.2 3e-05
xla:399210 rae1/gle2, Rae1; Rae1/Gle2 protein 45.1 5e-05
xla:446901 wipi1, MGC81027, atg18, wipi49; WD repeat domain, p... 37.4 0.011
xla:446709 MGC83946 protein 37.0 0.014
tpv:TP04_0670 hypothetical protein 35.4 0.039
mmu:52639 Wipi1, 4930533H01Rik, AW411817, D11Ertd498e, MGC3641... 34.7 0.070
cel:B0280.9 hypothetical protein; K14553 U3 small nucleolar RN... 33.9 0.11
mmu:237711 Eml6, 2900083P10Rik, C230094A16Rik, EMAP-6, MGC3893... 33.5 0.18
cel:F13H8.2 hypothetical protein; K14556 U3 small nucleolar RN... 33.1 0.25
cpv:cgd2_750 bub3'bub3-like protein with WD40 repeats' 32.7 0.27
dre:558292 Sterol regulatory element-binding protein cleavage-... 32.7 0.30
sce:YER082C UTP7, KRE31; Nucleolar protein, component of the s... 32.3 0.33
mmu:28000 Prpf19, AA617263, AL024362, D19Wsu55e, NMP200, PSO4,... 32.3 0.33
hsa:27339 PRPF19, NMP200, PRP19, PSO4, SNEV, UBOX4, hPSO4; PRP... 32.3 0.33
xla:380586 prpf19, MGC52755, nmp200; PRP19/PSO4 pre-mRNA proce... 32.0 0.43
hsa:55062 WIPI1, ATG18, ATG18A, FLJ10055, WIPI49; WD repeat do... 32.0 0.43
hsa:400954 EML6, DKFZp686G14111, FLJ16635, FLJ42562; echinoder... 32.0 0.46
cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing f... 32.0 0.47
xla:100127264 wdsub1, ubox6, wdsam1; WD repeat, sterile alpha ... 31.6 0.56
ath:AT3G10530 transducin family protein / WD-40 repeat family ... 31.6 0.60
ath:AT5G56130 transducin family protein / WD-40 repeat family ... 31.6 0.69
xla:398123 wdr1-a, MGC52751, aip1, wdr1b; WD repeat domain 1 31.6
hsa:161436 EML5, DKFZp781D1122, EMAP-2; echinoderm microtubule... 31.6 0.71
cel:Y45F10B.10 qui-1; QUInine non-avoider family member (qui-1) 31.2 0.81
dre:321544 prp19, NMP200, fb18f09, wu:fb18f09, zgc:56158; PRP1... 31.2 0.87
mmu:319670 Eml5, BC027154, C130068M19Rik; echinoderm microtubu... 31.2 0.87
mmu:217431 Nol10, Gm67, MGC113734; nucleolar protein 10; K1478... 30.8 1.0
hsa:22937 SCAP, KIAA0199; SREBF chaperone 30.8 1.0
tgo:TGME49_095680 periodic tryptophan protein PWP2, putative ;... 30.8 1.1
> tgo:TGME49_045470 mitotic checkpoint protein BUB3, putative
; K02180 cell cycle arrest protein BUB3
Length=332
Score = 125 bits (314), Expect = 4e-29, Method: Composition-based stats.
Identities = 56/80 (70%), Positives = 67/80 (83%), Gaps = 0/80 (0%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
YGTFATGGSDGGVS+WDG SKKRL R+P PTSV++LAFN SG LA+ VSY++E+GP P
Sbjct 249 YGTFATGGSDGGVSVWDGQSKKRLWRLPAFPTSVAALAFNPSGNQLAIGVSYLYEKGPIP 308
Query 86 GQPKPQIIVRAVREEDVRPK 105
P PQI+VR V++EDVRPK
Sbjct 309 TAPAPQIVVRLVKDEDVRPK 328
> ath:AT3G19590 WD-40 repeat family protein / mitotic checkpoint
protein, putative; K02180 cell cycle arrest protein BUB3
Length=340
Score = 87.4 bits (215), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 44/80 (55%), Positives = 58/80 (72%), Gaps = 1/80 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
YGTFATGG DG V+IWDG +KKRL + PTS+S+L+F+ G LLA+A SY FE G +
Sbjct 253 YGTFATGGCDGFVNIWDGNNKKRLYQYSKYPTSISALSFSRDGQLLAVASSYTFEEGEKS 312
Query 86 GQPKPQIIVRAVREEDVRPK 105
+P+ I VR+V E +V+PK
Sbjct 313 QEPEA-IFVRSVNEIEVKPK 331
> ath:AT1G49910 WD-40 repeat family protein / mitotic checkpoint
protein, putative; K02180 cell cycle arrest protein BUB3
Length=339
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 43/80 (53%), Positives = 58/80 (72%), Gaps = 1/80 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
YGTFA+GG DG V+IWDG +KKRL + PTS+++L+F+ G LLA+A SY FE G +P
Sbjct 252 YGTFASGGCDGFVNIWDGNNKKRLYQYSKYPTSIAALSFSRDGGLLAVASSYTFEEGDKP 311
Query 86 GQPKPQIIVRAVREEDVRPK 105
+P I VR+V E +V+PK
Sbjct 312 HEPD-AIFVRSVNEIEVKPK 330
> bbo:BBOV_II006880 18.m10036; WD domain/ mitotic checkpoint protein;
K02180 cell cycle arrest protein BUB3
Length=356
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/80 (50%), Positives = 58/80 (72%), Gaps = 1/80 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GTF TGG+DG V WDG+S+KRL R LPT+V+S++FN+SG LA+AVS MF+ QP
Sbjct 274 FGTFVTGGADGIVCAWDGISRKRLWRTTALPTAVASVSFNNSGEKLAIAVSDMFQVNGQP 333
Query 86 GQPKPQIIVRAVREEDVRPK 105
+P I+VR + ++ +P+
Sbjct 334 -TSQPSIMVRGISADECKPR 352
> mmu:12237 Bub3, AU019800, AU021329, AU043350, AW146323, C78067;
budding uninhibited by benzimidazoles 3 homolog (S. cerevisiae);
K02180 cell cycle arrest protein BUB3
Length=326
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 40/81 (49%), Positives = 56/81 (69%), Gaps = 1/81 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+ TFATGGSDG V+IWD +KKRLC+ PTS++SLAF++ GT LA+A SYM+E
Sbjct 246 HNTFATGGSDGFVNIWDPFNKKRLCQFHRYPTSIASLAFSNDGTTLAIASSYMYEMDDTE 305
Query 86 GQPKPQIIVRAVREEDVRPKT 106
P+ I +R V + + +PK+
Sbjct 306 -HPEDGIFIRQVTDAETKPKS 325
> hsa:9184 BUB3, BUB3L, hBUB3; budding uninhibited by benzimidazoles
3 homolog (yeast); K02180 cell cycle arrest protein BUB3
Length=326
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 40/81 (49%), Positives = 56/81 (69%), Gaps = 1/81 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+ TFATGGSDG V+IWD +KKRLC+ PTS++SLAF++ GT LA+A SYM+E
Sbjct 246 HNTFATGGSDGFVNIWDPFNKKRLCQFHRYPTSIASLAFSNDGTTLAIASSYMYEMDDTE 305
Query 86 GQPKPQIIVRAVREEDVRPKT 106
P+ I +R V + + +PK+
Sbjct 306 -HPEDGIFIRQVTDAETKPKS 325
> xla:399106 bub3, xbub3; budding uninhibited by benzimidazoles
3 homolog; K02180 cell cycle arrest protein BUB3
Length=330
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 39/80 (48%), Positives = 55/80 (68%), Gaps = 1/80 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+ TFATGGSDG V+IWD +KKRLC+ PTS++SLAF++ G+ LA+A SYM+E
Sbjct 252 HNTFATGGSDGFVNIWDPFNKKRLCQFHRYPTSIASLAFSNDGSTLAIAASYMYEMD-DI 310
Query 86 GQPKPQIIVRAVREEDVRPK 105
P+ I +R V + + +PK
Sbjct 311 DHPEDAIYIRQVTDAETKPK 330
> dre:403012 bub3, MGC101571, zgc:101571; BUB3 budding uninhibited
by benzimidazoles 3 homolog (yeast); K02180 cell cycle
arrest protein BUB3
Length=326
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 38/81 (46%), Positives = 56/81 (69%), Gaps = 1/81 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+ TFATGGSDG V+IWD +KKRLC+ P+S++SL+F++ G+LLA+A SYM E G
Sbjct 246 HNTFATGGSDGFVNIWDPFNKKRLCQFHRYPSSIASLSFSTDGSLLAIASSYMQELG-DV 304
Query 86 GQPKPQIIVRAVREEDVRPKT 106
P + +R V + + +PK+
Sbjct 305 SHPADAVFIRQVTDAETKPKS 325
> cel:Y54G9A.6 bub-3; yeast BUB homolog family member (bub-3);
K02180 cell cycle arrest protein BUB3
Length=343
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 54/81 (66%), Gaps = 1/81 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
YGTFATGG+DG V+IWD ++KR+ ++ TS+SSL+FN G+ LA+A SY +E+ P
Sbjct 263 YGTFATGGADGIVNIWDPFNRKRIIQLHKFETSISSLSFNEDGSQLAIATSYQYEKEIDP 322
Query 86 GQ-PKPQIIVRAVREEDVRPK 105
P I +R + + + RPK
Sbjct 323 SPLPNNSITIRHITDPESRPK 343
> tgo:TGME49_072350 poly(A)+ RNA export protein, putative ; K14298
mRNA export factor
Length=375
Score = 65.5 bits (158), Expect = 3e-11, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 53/83 (63%), Gaps = 3/83 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSG-TLLAMAVSYMFERGP- 83
+GTFATGG+DG + WD +++++L + SV+ + FN +G LLA AVSY + +GP
Sbjct 286 HGTFATGGADGSIVCWDKVNRQKLRAFDNMGNSVTDVKFNPTGNNLLAYAVSYDWSKGPD 345
Query 84 -QPGQPKPQIIVRAVREEDVRPK 105
Q Q+ V V++ED+RP+
Sbjct 346 QQELNKGHQVYVHMVKDEDIRPR 368
> tpv:TP02_0663 hypothetical protein; K02180 cell cycle arrest
protein BUB3
Length=302
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 49/75 (65%), Gaps = 1/75 (1%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GTF TGG DG + WDG+S+KRL + +V+S++FN SG LA+AVS +F+ P
Sbjct 222 FGTFVTGGGDGLLCGWDGISRKRLWKSSKFNGTVASVSFNHSGEKLAIAVSDVFQLNPHQ 281
Query 86 GQPKPQIIVRAVREE 100
Q P + ++ +++E
Sbjct 282 SQ-SPSLHLKHLKDE 295
> cpv:cgd6_4610 mRNA export protein ; K14298 mRNA export factor
Length=333
Score = 61.2 bits (147), Expect = 7e-10, Method: Composition-based stats.
Identities = 27/57 (47%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERG 82
YGTFATGGSDG ++ WD +K RL + +P V+ + F+ SG LLA ++SY + +G
Sbjct 247 YGTFATGGSDGAIAFWDKDNKSRLTIMKTMPAPVTDIKFSPSGKLLAYSLSYDWSKG 303
> ath:AT1G69400 transducin family protein / WD-40 repeat family
protein; K02180 cell cycle arrest protein BUB3
Length=314
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 24 CSYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGP 83
C GTF TG ++G V W+ S++RL +P S++SLAF+ +G LLA+A S+ ++
Sbjct 242 CGSGTFVTGDNEGYVISWNAKSRRRLNELPRYSNSIASLAFDHTGELLAIASSHTYQDAK 301
Query 84 QPGQPKPQIIVR 95
+ + PQ+ +
Sbjct 302 EK-EEAPQVFIH 312
> sce:YER107C GLE2, RAE1; Component of the Nup82 subcomplex of
the nuclear pore complex; required for polyadenylated RNA export
but not for protein import; homologous to S. pombe Rae1p;
K14298 mRNA export factor
Length=365
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 46/82 (56%), Gaps = 2/82 (2%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
YGTF T G DG + WD + RL P L S+ +FN +G++ A A+SY + +G
Sbjct 281 YGTFVTAGGDGTFNFWDKNQRHRLKGYPTLQASIPVCSFNRNGSVFAYALSYDWHQGHMG 340
Query 86 GQPK-PQII-VRAVREEDVRPK 105
+P P +I + A +E+V+ K
Sbjct 341 NRPDYPNVIRLHATTDEEVKEK 362
> tpv:TP01_0140 mRNA export protein; K14298 mRNA export factor
Length=359
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
YGTF +GG DG +IWD +K R+ L V + F S G LLA A SY + +G
Sbjct 269 YGTFVSGGGDGTFTIWDKDNKSRVKAFSNLGAPVVDVKFMSEGNLLAFATSYDWYKGLNH 328
Query 86 G---QPKPQIIVRAVREEDVRPK 105
I + ++EED++ K
Sbjct 329 SLITNTSKSIGIVKLKEEDIKSK 351
> bbo:BBOV_IV001150 21.m03073; mRNA export protein; K14298 mRNA
export factor
Length=359
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 3/84 (3%)
Query 25 SYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQ 84
++GTF TGG DG IWD ++ RL + + V + +S T+LA A SY + +G
Sbjct 267 NHGTFVTGGGDGNFVIWDKDNRSRLKQFNNVDAPVVDVKLHSDTTILAYATSYDWYKGYN 326
Query 85 PG---QPKPQIIVRAVREEDVRPK 105
+ + QI V +R ED +P+
Sbjct 327 QDLLMKTRRQIGVMQLRSEDFKPR 350
> ath:AT1G80670 transducin family protein / WD-40 repeat family
protein; K14298 mRNA export factor
Length=349
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 28/83 (33%), Positives = 43/83 (51%), Gaps = 3/83 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQ- 84
+GTFAT GSDG + WD SK+RL + + +FN G++ A A Y + +G +
Sbjct 258 HGTFATAGSDGAFNFWDKDSKQRLKAMSRCNQPIPCSSFNHDGSIYAYAACYDWSKGAEN 317
Query 85 --PGQPKPQIIVRAVREEDVRPK 105
P K I + +E +V+ K
Sbjct 318 HNPATAKSSIFLHLPQESEVKAK 340
> cel:F10G8.3 npp-17; Nuclear Pore complex Protein family member
(npp-17); K14298 mRNA export factor
Length=373
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 44/81 (54%), Gaps = 2/81 (2%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GT T GSDG S+WD ++ +L P P ++ +SSG L A+ Y + RG +
Sbjct 290 HGTLVTIGSDGRYSMWDKDARTKLKTSEPHPMPLTCCDVHSSGAFLVYALGYDWSRGHEG 349
Query 86 G-QPKPQIIVRAVREEDVRPK 105
QP +I++ ED++P+
Sbjct 350 NTQPGSKIVIHKCI-EDMKPR 369
> hsa:8480 RAE1, FLJ30608, MGC117333, MGC126076, MGC126077, MIG14,
MRNP41, Mnrp41, dJ481F12.3, dJ800J21.1; RAE1 RNA export
1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/82 (34%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GT AT GSDG S WD ++ +L L +S+ FN +G + A A SY + +G +
Sbjct 285 HGTLATVGSDGRFSFWDKDARTKLKTSEQLDQPISACCFNHNGNIFAYASSYDWSKGHEF 344
Query 86 GQPKPQ--IIVRAVREEDVRPK 105
P+ + I +R EE ++P+
Sbjct 345 YNPQKKNYIFLRNAAEE-LKPR 365
> xla:100049109 rae1, gle2, mig14, mnrp41, mrnp41; RAE1 RNA export
1 homolog; K14298 mRNA export factor
Length=368
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 46/82 (56%), Gaps = 3/82 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GT AT GSDG S WD ++ +L L +S+ +FN +G + A + SY + +G +
Sbjct 285 HGTLATVGSDGRFSFWDKDARTKLKTSEQLDQPISACSFNHNGNIFAYSSSYDWSKGHEF 344
Query 86 GQPKPQ--IIVRAVREEDVRPK 105
P+ + I +R EE ++P+
Sbjct 345 YNPQKKNYIFLRNAAEE-LKPR 365
> mmu:66679 Rae1, 3230401I12Rik, 41, D2Ertd342e, MNRP, MNRP41;
RAE1 RNA export 1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GT AT GSDG S WD ++ +L L +++ FN +G + A A SY + +G +
Sbjct 285 HGTLATVGSDGRFSFWDKDARTKLKTSEQLDQPIAACCFNHNGNIFAYASSYDWSKGHEF 344
Query 86 GQPKPQ--IIVRAVREEDVRPK 105
P+ + I +R EE ++P+
Sbjct 345 YNPQKKNYIFLRNAAEE-LKPR 365
> dre:393973 rae1, MGC56449, zgc:56449, zgc:77723; RAE1 RNA export
1 homolog (S. pombe); K14298 mRNA export factor
Length=368
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GT AT GSDG S WD ++ +L L +++ FN +G + A A SY + +G +
Sbjct 285 HGTLATVGSDGRFSFWDKDARTKLKTSEQLDQPITACCFNHNGNIFAYASSYDWSKGHEY 344
Query 86 GQPKPQ--IIVRAVREEDVRPK 105
P+ + I +R EE ++P+
Sbjct 345 YNPQKKNYIFLRNAAEE-LKPR 365
> xla:399210 rae1/gle2, Rae1; Rae1/Gle2 protein
Length=368
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQP 85
+GT AT GSDG S WD ++ +L L +++ FN +G + A + SY + +G +
Sbjct 285 HGTLATVGSDGRFSFWDKDARTKLKTSEQLDQPITACCFNHNGNIFAYSSSYDWSKGHEF 344
Query 86 GQPKPQ--IIVRAVREEDVRPK 105
P+ + I +R EE ++P+
Sbjct 345 YNPQKKNYIFLRNAAEE-LKPR 365
> xla:446901 wipi1, MGC81027, atg18, wipi49; WD repeat domain,
phosphoinositide interacting 1
Length=433
Score = 37.4 bits (85), Expect = 0.011, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 25 SYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
SY + S G VS++D K C +P + ++++AFNS+GT LA A
Sbjct 151 SYLAYPGSSSTGEVSLYDANCLKCECTIPAHDSPLAAIAFNSTGTKLASA 200
> xla:446709 MGC83946 protein
Length=433
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 25 SYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
SY + + S G VS++D S K C +P + ++++AFNS+GT LA A
Sbjct 151 SYLAYPSSSSSGEVSLYDANSLKCECTIPAHDSPLAAIAFNSTGTKLASA 200
> tpv:TP04_0670 hypothetical protein
Length=410
Score = 35.4 bits (80), Expect = 0.039, Method: Composition-based stats.
Identities = 21/46 (45%), Positives = 30/46 (65%), Gaps = 3/46 (6%)
Query 29 FATGGSDGGVSIWDGLSKKRLC--RVPPLPTSVSSLAFNSSGTLLA 72
ATGGSD V+++D + + +C P L VSSL+F+S+G LLA
Sbjct 265 LATGGSDHVVNLFD-VDSRMVCITTFPRLEGQVSSLSFSSNGALLA 309
> mmu:52639 Wipi1, 4930533H01Rik, AW411817, D11Ertd498e, MGC36416;
WD repeat domain, phosphoinositide interacting 1
Length=446
Score = 34.7 bits (78), Expect = 0.070, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 25 SYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
SY + S G + ++DG S K +C + +++++ FNSSG+ LA A
Sbjct 155 SYLAYPGSQSTGEIVLYDGNSLKTVCTIAAHEGTLAAITFNSSGSKLASA 204
> cel:B0280.9 hypothetical protein; K14553 U3 small nucleolar
RNA-associated protein 18
Length=429
Score = 33.9 bits (76), Expect = 0.11, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 33/60 (55%), Gaps = 7/60 (11%)
Query 21 LRICSYGT-FATGGSDGGVSIWDG------LSKKRLCRVPPLPTSVSSLAFNSSGTLLAM 73
L I +G FATG G V+++ G + + L V L T+VSS+AFNS L+A+
Sbjct 305 LAISQHGDYFATGSDTGIVNVYSGNDCRNSTNPRPLFNVSNLVTAVSSIAFNSDAQLMAI 364
> mmu:237711 Eml6, 2900083P10Rik, C230094A16Rik, EMAP-6, MGC38936;
echinoderm microtubule associated protein like 6
Length=1958
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 1 WNGKWLFTLIREFNSFFLVALRICSYGTFATGGSDGGVSIWD 42
W G L I+ +S + +L C G FATGG DG + +WD
Sbjct 223 WKGLTLVRTIQGAHSAGIFSLYACEEG-FATGGRDGCIRLWD 263
> cel:F13H8.2 hypothetical protein; K14556 U3 small nucleolar
RNA-associated protein 12
Length=929
Score = 33.1 bits (74), Expect = 0.25, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAF 64
FATGG DG + +WD L+++ + R+ SV+ + F
Sbjct 118 FATGGKDGVIVLWDILAERGMFRLHGHKESVTQMKF 153
> cpv:cgd2_750 bub3'bub3-like protein with WD40 repeats'
Length=422
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 14/23 (60%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 29 FATGGSDGGVSIWDGLSKKRLCR 51
ATGGSD V +WD +KKRL R
Sbjct 319 LATGGSDASVFLWDTSAKKRLWR 341
> dre:558292 Sterol regulatory element-binding protein cleavage-activating
protein-like
Length=1245
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query 6 LFTLIREFNSFFLVALRICSYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFN 65
LFTL + +S + A+ I A+GG DG + +WD L+ R+ V V+SL
Sbjct 1078 LFTL--QGHSGGITAIYIDQTMVLASGGQDGAICVWDVLTGSRVSHVYGHRGDVTSLVCT 1135
Query 66 SS 67
+S
Sbjct 1136 TS 1137
> sce:YER082C UTP7, KRE31; Nucleolar protein, component of the
small subunit (SSU) processome containing the U3 snoRNA that
is involved in processing of pre-18S rRNA; K14768 U3 small
nucleolar RNA-associated protein 7
Length=554
Score = 32.3 bits (72), Expect = 0.33, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 32/55 (58%), Gaps = 7/55 (12%)
Query 30 ATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFERGPQ 84
AT G+D + IWD + K+L V LPT ++++ + +G LLA++ RGP
Sbjct 290 ATTGADRSMKIWDIRNFKQLHSVESLPTPGTNVSISDTG-LLALS------RGPH 337
> mmu:28000 Prpf19, AA617263, AL024362, D19Wsu55e, NMP200, PSO4,
Prp19, Snev; PRP19/PSO4 pre-mRNA processing factor 19 homolog
(S. cerevisiae); K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=504
Score = 32.3 bits (72), Expect = 0.33, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
F TG D + IWD + + P ++S+AF+ +G LA A
Sbjct 365 FGTGTMDSQIKIWDLKERTNVANFPGHSGPITSIAFSENGYYLATA 410
> hsa:27339 PRPF19, NMP200, PRP19, PSO4, SNEV, UBOX4, hPSO4; PRP19/PSO4
pre-mRNA processing factor 19 homolog (S. cerevisiae);
K10599 pre-mRNA-processing factor 19 [EC:6.3.2.19]
Length=504
Score = 32.3 bits (72), Expect = 0.33, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
F TG D + IWD + + P ++S+AF+ +G LA A
Sbjct 365 FGTGTMDSQIKIWDLKERTNVANFPGHSGPITSIAFSENGYYLATA 410
> xla:380586 prpf19, MGC52755, nmp200; PRP19/PSO4 pre-mRNA processing
factor 19 homolog; K10599 pre-mRNA-processing factor
19 [EC:6.3.2.19]
Length=504
Score = 32.0 bits (71), Expect = 0.43, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
F TG D + IWD + + P VS +AF+ +G LA A
Sbjct 365 FGTGTVDSQIKIWDLKERSNVANFPGHSGPVSCIAFSENGYYLATA 410
> hsa:55062 WIPI1, ATG18, ATG18A, FLJ10055, WIPI49; WD repeat
domain, phosphoinositide interacting 1
Length=446
Score = 32.0 bits (71), Expect = 0.43, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 25 SYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
SY + + G + ++DG S K +C + +++++ FN+SG+ LA A
Sbjct 155 SYLAYPGSLTSGEIVLYDGNSLKTVCTIAAHEGTLAAITFNASGSKLASA 204
> hsa:400954 EML6, DKFZp686G14111, FLJ16635, FLJ42562; echinoderm
microtubule associated protein like 6
Length=1958
Score = 32.0 bits (71), Expect = 0.46, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 1 WNGKWLFTLIREFNSFFLVALRICSYGTFATGGSDGGVSIWD 42
W G L I+ +S + ++ C G FATGG DG + +WD
Sbjct 223 WKGLNLVRTIQGAHSAGIFSMYACEEG-FATGGRDGCIRLWD 263
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLL 71
F TGG DG V +WD + ++ L S+L+ +S G LL
Sbjct 916 FVTGGKDGIVELWDDMFERCL---KTYAIKRSALSTSSKGLLL 955
> cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=492
Score = 32.0 bits (71), Expect = 0.47, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLA 72
F TG +D V IWD ++ P +V S+AF+ +G LA
Sbjct 353 FGTGAADAVVKIWDLKNQTVAAAFPGHTAAVRSIAFSENGYYLA 396
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVP-PLPTSVSSLAFNSSGTLLAMA 74
ATG DG V +WD K L ++SL+F+ +GT L +
Sbjct 395 LATGSEDGEVKLWDLRKLKNLKTFANEEKQPINSLSFDMTGTFLGIG 441
> xla:100127264 wdsub1, ubox6, wdsam1; WD repeat, sterile alpha
motif and U-box domain containing 1
Length=460
Score = 31.6 bits (70), Expect = 0.56, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 4/60 (6%)
Query 21 LRICSYGT----FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVS 76
+R+C Y + ATGG+DG + +W+ K S+ + AF+ G LL S
Sbjct 100 VRVCRYSSNSNYLATGGADGSIVLWNVQQMKFYRSATVKDGSIVACAFSPHGNLLITGSS 159
> ath:AT3G10530 transducin family protein / WD-40 repeat family
protein; K14768 U3 small nucleolar RNA-associated protein
7
Length=536
Score = 31.6 bits (70), Expect = 0.60, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 32 GGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
G S G V++W S+ L ++ P VSS+AF+ +G L+A +
Sbjct 257 GHSGGTVTMWKPTSQAPLVQMQCHPGPVSSVAFHPNGHLMATS 299
> ath:AT5G56130 transducin family protein / WD-40 repeat family
protein; K12880 THO complex subunit 3
Length=315
Score = 31.6 bits (70), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 32/71 (45%), Gaps = 2/71 (2%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMAVSYMFER--GPQPG 86
FA G +D VS+WD L L V +++FN SG +A A +F Q G
Sbjct 206 FAVGSADSLVSLWDISDMLCLRTFTKLEWPVRTISFNYSGEYIASASEDLFIDIANVQTG 265
Query 87 QPKPQIIVRAV 97
+ QI RA
Sbjct 266 RTVHQIPCRAA 276
> xla:398123 wdr1-a, MGC52751, aip1, wdr1b; WD repeat domain 1
Length=608
Score = 31.6 bits (70), Expect = 0.70, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 27 GTFATGGSDGGVSIWD--GLSKKRLCRVPPLPTSVSSLAFNSSGTLLAM 73
GT A GG+DG V ++ G S K + P +V+ LA++ G LA+
Sbjct 458 GTVAVGGADGKVHLYSIQGNSLKDEGKTLPAKGAVTDLAYSHDGAFLAV 506
> hsa:161436 EML5, DKFZp781D1122, EMAP-2; echinoderm microtubule
associated protein like 5
Length=1977
Score = 31.6 bits (70), Expect = 0.71, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 1 WNGKWLFTLIREFNSFFLVALRICSYGTFATGGSDGGVSIWD 42
W G L I+ ++ + ++ C G FATGG DG + +WD
Sbjct 223 WKGINLIRTIQGAHAAGIFSMNACEEG-FATGGRDGCIRLWD 263
> cel:Y45F10B.10 qui-1; QUInine non-avoider family member (qui-1)
Length=1592
Score = 31.2 bits (69), Expect = 0.81, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 3/69 (4%)
Query 9 LIREFNSFF--LVALRICSYGTF-ATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFN 65
++R FN +V+L++ S F TG D V +WD + K + R+ L VS+LA
Sbjct 987 VMRSFNDHTGSVVSLQLTSNNQFLITGSGDFVVQMWDVTNGKCISRMGGLMAPVSTLAIT 1046
Query 66 SSGTLLAMA 74
S+ + +A
Sbjct 1047 SNDAFVVVA 1055
> dre:321544 prp19, NMP200, fb18f09, wu:fb18f09, zgc:56158; PRP19/PSO4
homolog (S. cerevisiae); K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=505
Score = 31.2 bits (69), Expect = 0.87, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLA 72
F TG D + IWD + + P V+++AF+ +G LA
Sbjct 366 FGTGTGDSQIKIWDLKERTNVANFPGHSGPVTAIAFSENGYYLA 409
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 29 FATGGSDGGVSIWDGLSKKRLCRVPPLPTS--VSSLAFNSSGTLLAMAVS 76
ATG D + +WD L K + + L + V SL F+ SGT LA+ S
Sbjct 408 LATGAQDSSLKLWD-LRKLKNFKTITLDNNYEVKSLVFDQSGTYLAVGGS 456
> mmu:319670 Eml5, BC027154, C130068M19Rik; echinoderm microtubule
associated protein like 5
Length=1977
Score = 31.2 bits (69), Expect = 0.87, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 1 WNGKWLFTLIREFNSFFLVALRICSYGTFATGGSDGGVSIWD 42
W G L I+ ++ + ++ C G FATGG DG + +WD
Sbjct 223 WKGINLIRTIQGAHTAGIFSMNSCEEG-FATGGRDGCIRLWD 263
> mmu:217431 Nol10, Gm67, MGC113734; nucleolar protein 10; K14788
ribosome biogenesis protein ENP2
Length=687
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 17/63 (26%)
Query 26 YGTFATGGSDGGVSIWDGLSKKRL-------------CRVPPLPTSVSSLAFNSSGTLLA 72
+G FATG +G V WD +KR+ + LPT +S+L FN + L+
Sbjct 188 HGLFATGTIEGRVECWDPRVRKRVGVLDCALNSVTADSEINSLPT-ISALKFNGA---LS 243
Query 73 MAV 75
MAV
Sbjct 244 MAV 246
> hsa:22937 SCAP, KIAA0199; SREBF chaperone
Length=1279
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 31/62 (50%), Gaps = 2/62 (3%)
Query 6 LFTLIREFNSFFLVALRICSYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFN 65
LFTL + +S + + I A+GG DG + +WD L+ R+ V V+SL
Sbjct 1112 LFTL--QGHSGAITTVYIDQTMVLASGGQDGAICLWDVLTGSRVSHVFAHRGDVTSLTCT 1169
Query 66 SS 67
+S
Sbjct 1170 TS 1171
> tgo:TGME49_095680 periodic tryptophan protein PWP2, putative
; K14558 periodic tryptophan protein 2
Length=1266
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 25 SYGTFATGGSDGGVSIWDGLSKKRLCRVPPLPTSVSSLAFNSSGTLLAMA 74
S G ATGG+DG V ++D + C +V++L F S G + A
Sbjct 639 SRGIVATGGTDGRVKLFDAETGFCFCSFADHAAAVTALVFASGGNAVFTA 688
Lambda K H
0.322 0.138 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012750684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40