bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3138_orf1
Length=170
Score E
Sequences producing significant alignments: (Bits) Value
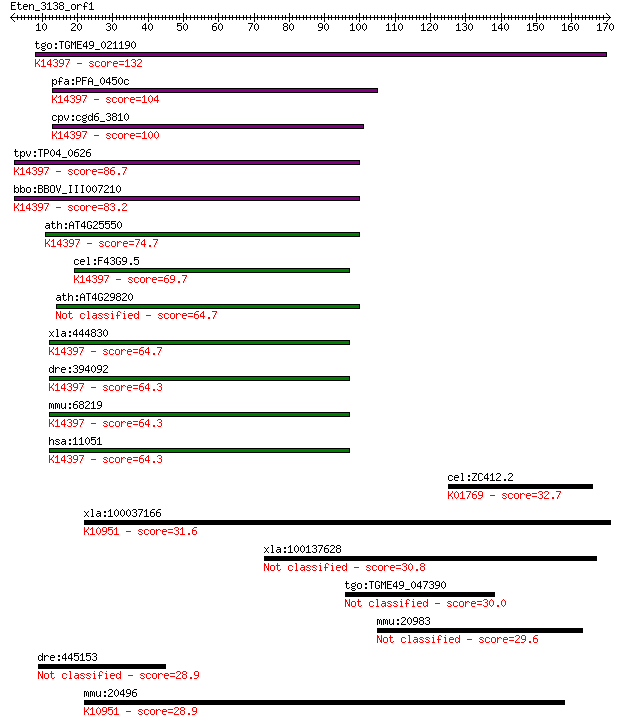
tgo:TGME49_021190 mRNA cleavage factor-like protein, putative ... 132 4e-31
pfa:PFA_0450c mRNA cleavage factor-like protein, putative; K14... 104 1e-22
cpv:cgd6_3810 NUDIX domain protein; mRNA cleavage factor-like ... 100 2e-21
tpv:TP04_0626 mRNA cleavage factor protein; K14397 cleavage an... 86.7 4e-17
bbo:BBOV_III007210 17.m07632; hypothetical protein; K14397 cle... 83.2 4e-16
ath:AT4G25550 protein binding; K14397 cleavage and polyadenyla... 74.7 2e-13
cel:F43G9.5 hypothetical protein; K14397 cleavage and polyaden... 69.7 4e-12
ath:AT4G29820 CFIM-25 64.7 1e-10
xla:444830 nudt21, MGC84447, cpsf5; nudix (nucleoside diphosph... 64.7 2e-10
dre:394092 cpsf5, MGC63966, zgc:63966; cleavage and polyadenyl... 64.3 2e-10
mmu:68219 Nudt21, 25kDa, 3110048P04Rik, 5730530J16Rik, AU01486... 64.3 2e-10
hsa:11051 NUDT21, CFIM25, CPSF5, DKFZp686H1588; nudix (nucleos... 64.3 2e-10
cel:ZC412.2 gcy-14; Guanylyl CYclase family member (gcy-14); K... 32.7 0.63
xla:100037166 nkcc1; Na-K-2Cl cotransporter 1; K10951 solute c... 31.6 1.3
xla:100137628 hypothetical protein LOC100137628 30.8 2.5
tgo:TGME49_047390 ATPase, AAA family domain-containing protein... 30.0 4.2
mmu:20983 Syt4, SytIV; synaptotagmin IV 29.6 5.6
dre:445153 im:7149072; si:ch211-175f11.2 28.9 8.0
mmu:20496 Slc12a2, 9330166H04Rik, Nkcc1, mBSC2, sy-ns; solute ... 28.9 8.8
> tgo:TGME49_021190 mRNA cleavage factor-like protein, putative
; K14397 cleavage and polyadenylation specificity factor subunit
5
Length=414
Score = 132 bits (333), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 74/167 (44%), Positives = 103/167 (61%), Gaps = 11/167 (6%)
Query 8 TDGPDLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCL 67
++G EVGEFLGEWWR EFD++ P+LPPHVTRPKER+R++QVQLP +CSFR+P F L
Sbjct 253 SEGVAAEVGEFLGEWWRVEFDEEPQPFLPPHVTRPKERIRLYQVQLPPKCSFRLPPAFSL 312
Query 68 TVVPIFE-LSPQRVGLAIGGLSHLIARFSIRLMVPTLDKYEADTSLEH----KGEAGLDD 122
+P+F+ L P+ G+A+ GL H+++RF RL+ P+LD + + +L + E ++
Sbjct 313 AALPLFDLLRPEIHGVALSGLPHVVSRFRFRLLSPSLDGFVSSANLRRPRGGQQEGDAEE 372
Query 123 LGTSFDIHELEEQPDVAGGHSEAKPALENDGELEMLPPELQHLAELE 169
L + + ELE G G E LPPEL LAE E
Sbjct 373 LSGEY-LAELERMNTSQDGELS-----HIHGATEELPPELAALAEEE 413
> pfa:PFA_0450c mRNA cleavage factor-like protein, putative; K14397
cleavage and polyadenylation specificity factor subunit
5
Length=232
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 44/92 (47%), Positives = 65/92 (70%), Gaps = 2/92 (2%)
Query 13 LEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPI 72
+E+G+FLGEWWR +F+ +PYLP H++RPKE +R++QV L +C F +P GF L +P+
Sbjct 140 VEIGDFLGEWWRTQFNSVFLPYLPAHISRPKEYIRLYQVILSPKCIFHLPPGFTLKAIPL 199
Query 73 FELSPQRVGLAIGGLSHLIARFSIRLMVPTLD 104
F+L+ GLAI GLS +++RF + MV D
Sbjct 200 FDLN--NCGLAISGLSSILSRFKLHCMVQEQD 229
> cpv:cgd6_3810 NUDIX domain protein; mRNA cleavage factor-like
protein Im like, plant+animal group ; K14397 cleavage and
polyadenylation specificity factor subunit 5
Length=277
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 44/88 (50%), Positives = 61/88 (69%), Gaps = 0/88 (0%)
Query 13 LEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPI 72
+EVGE+LG WWR EF+ +PYLPPH TRPKE +R++QV LP + F++P L +P+
Sbjct 186 VEVGEYLGTWWRTEFNYSPLPYLPPHSTRPKETIRIYQVILPPKLLFKLPKHHVLKSLPL 245
Query 73 FELSPQRVGLAIGGLSHLIARFSIRLMV 100
F+L P G+A G + LI+RF I+ MV
Sbjct 246 FDLDPNIFGIACGSIPQLISRFQIQSMV 273
> tpv:TP04_0626 mRNA cleavage factor protein; K14397 cleavage
and polyadenylation specificity factor subunit 5
Length=226
Score = 86.7 bits (213), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 38/100 (38%), Positives = 61/100 (61%), Gaps = 2/100 (2%)
Query 2 HKKQLSTDGP--DLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSF 59
HK QL+ ++VGE L ++WR +F+ + +PYLP H RPKE++ ++QV L C
Sbjct 124 HKHQLNIKETIETIQVGELLADFWRCDFNTEPLPYLPLHTNRPKEKISLYQVVLQESCKI 183
Query 60 RIPVGFCLTVVPIFELSPQRVGLAIGGLSHLIARFSIRLM 99
+P G+ L VP+++ GL++G L HL++RF I +
Sbjct 184 SVPKGYSLKFVPLYDFYNPEFGLSLGSLPHLLSRFKISYL 223
> bbo:BBOV_III007210 17.m07632; hypothetical protein; K14397 cleavage
and polyadenylation specificity factor subunit 5
Length=357
Score = 83.2 bits (204), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 59/98 (60%), Gaps = 0/98 (0%)
Query 2 HKKQLSTDGPDLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRI 61
H+ + + + VGEF+GE+WR E+D D++PYLP H+ RP+E++ ++QV L +CSF
Sbjct 121 HQLNVRANVDTIIVGEFMGEFWRAEYDSDVLPYLPLHINRPREKILIYQVTLREQCSFIA 180
Query 62 PVGFCLTVVPIFELSPQRVGLAIGGLSHLIARFSIRLM 99
P + + + E +AI L HL+ RF++ M
Sbjct 181 PGDMHIEPMALHEFYCAEQSVAISALPHLLTRFNLSFM 218
> ath:AT4G25550 protein binding; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=200
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 51/91 (56%), Gaps = 2/91 (2%)
Query 11 PDLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVV 70
PD VGE + WWR F+ + PY PPH+T+PKE R++ V L + F +P L V
Sbjct 108 PDWTVGECVATWWRPNFETMMYPYCPPHITKPKECKRLYIVHLSEKEYFAVPKNLKLLAV 167
Query 71 PIFEL--SPQRVGLAIGGLSHLIARFSIRLM 99
P+FEL + QR G I + ++RF ++
Sbjct 168 PLFELYDNVQRYGPVISTIPQQLSRFHFNMI 198
> cel:F43G9.5 hypothetical protein; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=227
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 45/80 (56%), Gaps = 2/80 (2%)
Query 19 LGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIFEL--S 76
+G WWR FD PY+P HVT+PKE ++ VQLP + +F +P F L P+FEL +
Sbjct 142 IGNWWRPNFDPPRYPYIPAHVTKPKEHTKLLLVQLPSKSTFCVPKNFKLVAAPLFELYDN 201
Query 77 PQRVGLAIGGLSHLIARFSI 96
G I L ++RF+
Sbjct 202 AAAYGPLISSLPTTLSRFNF 221
> ath:AT4G29820 CFIM-25
Length=222
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 51/88 (57%), Gaps = 2/88 (2%)
Query 14 EVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIF 73
EVGE +G WWR F+ + P+LPP++ PKE ++ V+LP F +P F L VP+
Sbjct 133 EVGECIGMWWRPNFETLMYPFLPPNIKHPKECTKLFLVRLPVHQQFVVPKNFKLLAVPLC 192
Query 74 EL--SPQRVGLAIGGLSHLIARFSIRLM 99
+L + + G + + L+++FS +M
Sbjct 193 QLHENEKTYGPIMSQIPKLLSKFSFNMM 220
> xla:444830 nudt21, MGC84447, cpsf5; nudix (nucleoside diphosphate
linked moiety X)-type motif 21; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=227
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 46/87 (52%), Gaps = 2/87 (2%)
Query 12 DLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVP 71
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F +P + L P
Sbjct 138 DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKNYKLVAAP 197
Query 72 IFELSPQRVGLA--IGGLSHLIARFSI 96
+FEL G I L L++RF+
Sbjct 198 LFELYDNAPGYGPIISSLPQLLSRFNF 224
> dre:394092 cpsf5, MGC63966, zgc:63966; cleavage and polyadenylation
specific factor 5; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=228
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 46/87 (52%), Gaps = 2/87 (2%)
Query 12 DLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVP 71
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F +P + L P
Sbjct 139 DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKNYKLVAAP 198
Query 72 IFELSPQRVGLA--IGGLSHLIARFSI 96
+FEL G I L L++RF+
Sbjct 199 LFELYDNAPGYGPIISSLPQLLSRFNF 225
> mmu:68219 Nudt21, 25kDa, 3110048P04Rik, 5730530J16Rik, AU014860,
AW549947, Cpsf5; nudix (nucleoside diphosphate linked moiety
X)-type motif 21; K14397 cleavage and polyadenylation
specificity factor subunit 5
Length=227
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 46/87 (52%), Gaps = 2/87 (2%)
Query 12 DLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVP 71
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F +P + L P
Sbjct 138 DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKNYKLVAAP 197
Query 72 IFELSPQRVGLA--IGGLSHLIARFSI 96
+FEL G I L L++RF+
Sbjct 198 LFELYDNAPGYGPIISSLPQLLSRFNF 224
> hsa:11051 NUDT21, CFIM25, CPSF5, DKFZp686H1588; nudix (nucleoside
diphosphate linked moiety X)-type motif 21; K14397 cleavage
and polyadenylation specificity factor subunit 5
Length=227
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 46/87 (52%), Gaps = 2/87 (2%)
Query 12 DLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVP 71
D + + +G WWR F+ PY+P H+T+PKE ++ VQL + F +P + L P
Sbjct 138 DWVIDDCIGNWWRPNFEPPQYPYIPAHITKPKEHKKLFLVQLQEKALFAVPKNYKLVAAP 197
Query 72 IFELSPQRVGLA--IGGLSHLIARFSI 96
+FEL G I L L++RF+
Sbjct 198 LFELYDNAPGYGPIISSLPQLLSRFNF 224
> cel:ZC412.2 gcy-14; Guanylyl CYclase family member (gcy-14);
K01769 guanylate cyclase, other [EC:4.6.1.2]
Length=1170
Score = 32.7 bits (73), Expect = 0.63, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 7/47 (14%)
Query 125 TSFDIHELEEQPDV------AGGHSEAKPALENDGELEMLPPELQHL 165
++FD+ +E+PDV GGH+ +P+L+ GE + P L HL
Sbjct 792 SAFDLENRKEKPDVIIYQVKKGGHNPMRPSLDT-GETVEINPALLHL 837
> xla:100037166 nkcc1; Na-K-2Cl cotransporter 1; K10951 solute
carrier family 12 (sodium/potassium/chloride transporter),
member 2
Length=1158
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 34/152 (22%), Positives = 63/152 (41%), Gaps = 27/152 (17%)
Query 22 WWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIFELSPQRVG 81
WW FDD + L P++ K++ R C R+ +G + R+
Sbjct 973 WWL--FDDGGLTLLIPYLITTKKKWR--------DCKIRVFIGGKI----------NRID 1012
Query 82 LAIGGLSHLIARFSIR---LMVPTLDKYEADTSLEHKGEAGLDDLGTSFDIHELEEQPDV 138
++ L+++F I +MV + +T + + A +++ F +HE E++ +V
Sbjct 1013 HDRRAMATLLSKFRIDFSDIMVLG----DINTKPKKENVAAFEEMIEPFRLHEDEKEQEV 1068
Query 139 AGGHSEAKPALENDGELEMLPPELQHLAELEE 170
A E +P D ELE+ + L E
Sbjct 1069 ADKMKEEEPWRITDNELELYKTKTHRQIRLNE 1100
> xla:100137628 hypothetical protein LOC100137628
Length=544
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 50/119 (42%), Gaps = 29/119 (24%)
Query 73 FELSPQRVGLAIGGLSHLIARFSIRLMVPTLDKY-----EADTSLEHKGEAGLDDLGTSF 127
++ P+ + L+HL + ++ + Y E+D+S EA ++L F
Sbjct 184 YQKQPEVKSTSYSRLNHLSSPYAYSTPLFKKASYRNLRKESDSS----NEASFEELSPCF 239
Query 128 DIHELEEQPDVAGGHSEAK-----------------PALENDGELEMLPP---ELQHLA 166
+L E+P AGG + K P LE+D E LPP E++HLA
Sbjct 240 SKEDLHEEPSHAGGMLKEKIHSLGQFIPTTRVLPLNPELESDEEFLSLPPHLKEIEHLA 298
> tgo:TGME49_047390 ATPase, AAA family domain-containing protein
(EC:2.7.11.1 4.1.1.70 3.4.21.72)
Length=3668
Score = 30.0 bits (66), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 96 IRLMVPTLDKYEADTSLEHKGEAGLDDLGTSFDIHELEEQPD 137
RL D EA+ S+E +GEA DD G S +EQ D
Sbjct 2689 ARLERQDADAREAEASMEERGEADRDDEGGSTSTCSGKEQED 2730
> mmu:20983 Syt4, SytIV; synaptotagmin IV
Length=425
Score = 29.6 bits (65), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 4/58 (6%)
Query 105 KYEADTSLEHKGEAGLDDLGTSFDIHELEEQPDVAGGHSEAKPALENDGELEMLPPEL 162
K+ D E KG+A L +L D+ E+ D+ G +A P + +LE + P+L
Sbjct 72 KFGGDDKSEVKGKAALPNLSLHLDL----EKRDLNGNFPKANPKAGSSSDLENVTPKL 125
> dre:445153 im:7149072; si:ch211-175f11.2
Length=1741
Score = 28.9 bits (63), Expect = 8.0, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 18/36 (50%), Gaps = 1/36 (2%)
Query 9 DGPDLEVGEFLGEWWRGEFDDDLVPYLPPHVTRPKE 44
+G + V E GEWWRG D P + +PKE
Sbjct 1049 EGDTILVSEREGEWWRGSIGDR-AGLFPSNYVKPKE 1083
> mmu:20496 Slc12a2, 9330166H04Rik, Nkcc1, mBSC2, sy-ns; solute
carrier family 12, member 2; K10951 solute carrier family
12 (sodium/potassium/chloride transporter), member 2
Length=1206
Score = 28.9 bits (63), Expect = 8.8, Method: Composition-based stats.
Identities = 30/139 (21%), Positives = 59/139 (42%), Gaps = 27/139 (19%)
Query 22 WWRGEFDDDLVPYLPPHVTRPKERVRVHQVQLPHRCSFRIPVGFCLTVVPIFELSPQRVG 81
WW FDD + L P++ K++ + C R+ +G + R+
Sbjct 1021 WWL--FDDGGLTLLIPYLLTTKKKWK--------DCKIRVFIGGKI----------NRID 1060
Query 82 LAIGGLSHLIARFSIR---LMVPTLDKYEADTSLEHKGEAGLDDLGTSFDIHELEEQPDV 138
++ L+++F I +MV + +T + + DD+ + +HE +++ D+
Sbjct 1061 HDRRAMATLLSKFRIDFSDIMVLG----DINTKPKKENIIAFDDMIEPYRLHEDDKEQDI 1116
Query 139 AGGHSEAKPALENDGELEM 157
A E +P D ELE+
Sbjct 1117 ADKMKEDEPWRITDNELEL 1135
Lambda K H
0.318 0.140 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4287611064
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40