bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3134_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
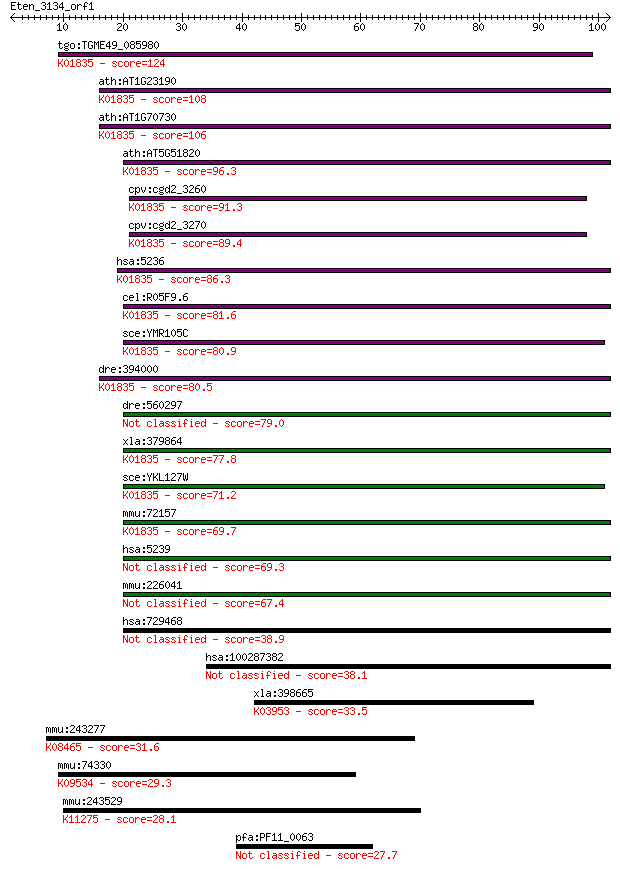
tgo:TGME49_085980 phosphoglucomutase/parafusin related protein... 124 7e-29
ath:AT1G23190 phosphoglucomutase, cytoplasmic, putative / gluc... 108 5e-24
ath:AT1G70730 phosphoglucomutase, cytoplasmic, putative / gluc... 106 2e-23
ath:AT5G51820 PGM; PGM (PHOSPHOGLUCOMUTASE); phosphoglucomutas... 96.3 2e-20
cpv:cgd2_3260 phosphoglucomutase, tandemly duplicated gene (EC... 91.3 6e-19
cpv:cgd2_3270 phosphoglucomutase, tandemly duplicated gene (EC... 89.4 2e-18
hsa:5236 PGM1, GSD14; phosphoglucomutase 1 (EC:5.4.2.2); K0183... 86.3 2e-17
cel:R05F9.6 hypothetical protein; K01835 phosphoglucomutase [E... 81.6 6e-16
sce:YMR105C PGM2, GAL5; Pgm2p (EC:5.4.2.2); K01835 phosphogluc... 80.9 9e-16
dre:394000 pgm1, MGC63718, zgc:63718; phosphoglucomutase 1 (EC... 80.5 1e-15
dre:560297 novel protein similar to vertebrate phosphoglucomut... 79.0 4e-15
xla:379864 pgm1, MGC53760, pgm2; phosphoglucomutase 1 (EC:5.4.... 77.8 7e-15
sce:YKL127W PGM1; Pgm1p (EC:5.4.2.2); K01835 phosphoglucomutas... 71.2 7e-13
mmu:72157 Pgm2, 2610020G18Rik, AA407108, AI098105, Pgm-2, Pgm1... 69.7 2e-12
hsa:5239 PGM5, PGMRP; phosphoglucomutase 5 69.3
mmu:226041 Pgm5, 4833423B07, 9530034F03Rik, D830025G17, acicul... 67.4 1e-11
hsa:729468 putative PGM5-like protein 1-like 38.9 0.004
hsa:100287382 putative PGM5-like protein 1-like 38.1 0.008
xla:398665 ndufa9, MGC64316, ndufs2l; NADH dehydrogenase (ubiq... 33.5 0.17
mmu:243277 Gpr133, E230012M21Rik; G protein-coupled receptor 1... 31.6 0.71
mmu:74330 Dnajc14, 5730551F12Rik, DNAJ, DRIP78, HDJ3, LIP6; Dn... 29.3 2.8
mmu:243529 H1fx, Gm461, H1X; H1 histone family, member X; K112... 28.1 6.4
pfa:PF11_0063 conserved Plasmodium protein 27.7 9.3
> tgo:TGME49_085980 phosphoglucomutase/parafusin related protein
1, putative (EC:5.4.2.2); K01835 phosphoglucomutase [EC:5.4.2.2]
Length=637
Score = 124 bits (311), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 55/90 (61%), Positives = 72/90 (80%), Gaps = 0/90 (0%)
Query 9 GVPPESASVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTML 68
G+P + R TT ++ QKPGTSGLR+KT+ FMQ YL+N+ Q+VF+CLPE E +GGT+L
Sbjct 25 GLPADQILSRPTTAYQDQKPGTSGLRKKTKVFMQKDYLANFAQSVFNCLPEEEKKGGTLL 84
Query 69 VAGDGRYFCSDAIKKIVAIAAGNGVGRVWI 98
V+GDGR+F +AI +I +IAAGNGVGRVWI
Sbjct 85 VSGDGRFFSHEAIYEICSIAAGNGVGRVWI 114
> ath:AT1G23190 phosphoglucomutase, cytoplasmic, putative / glucose
phosphomutase, putative (EC:5.4.2.2); K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=583
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 46/86 (53%), Positives = 65/86 (75%), Gaps = 0/86 (0%)
Query 16 SVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRY 75
S +T+P +GQKPGTSGLR+K + F QP YL N+VQA F+ L +V+G T++V+GDGRY
Sbjct 6 STVSTSPIDGQKPGTSGLRKKVKVFKQPNYLENFVQATFNALTAEKVKGATLVVSGDGRY 65
Query 76 FCSDAIKKIVAIAAGNGVGRVWIAKD 101
+ DA++ I+ +AA NGV RVW+ K+
Sbjct 66 YSKDAVQIIIKMAAANGVRRVWVGKN 91
> ath:AT1G70730 phosphoglucomutase, cytoplasmic, putative / glucose
phosphomutase, putative; K01835 phosphoglucomutase [EC:5.4.2.2]
Length=605
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 46/86 (53%), Positives = 65/86 (75%), Gaps = 0/86 (0%)
Query 16 SVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRY 75
S+ +T+P +GQKPGTSGLR+K + F QP YL N+VQA F+ L +V+G T++V+GDGRY
Sbjct 27 SLVSTSPIDGQKPGTSGLRKKVKVFKQPNYLENFVQATFNALTTEKVKGATLVVSGDGRY 86
Query 76 FCSDAIKKIVAIAAGNGVGRVWIAKD 101
+ AI+ IV +AA NGV RVW+ ++
Sbjct 87 YSEQAIQIIVKMAAANGVRRVWVGQN 112
> ath:AT5G51820 PGM; PGM (PHOSPHOGLUCOMUTASE); phosphoglucomutase
(EC:5.4.2.2); K01835 phosphoglucomutase [EC:5.4.2.2]
Length=623
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 41/82 (50%), Positives = 61/82 (74%), Gaps = 0/82 (0%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSD 79
T P EGQK GTSGLR+K + FM+ YL+N++QA+F+ LP + + T+++ GDGRYF +
Sbjct 74 TKPIEGQKTGTSGLRKKVKVFMEDNYLANWIQALFNSLPLEDYKNATLVLGGDGRYFNKE 133
Query 80 AIKKIVAIAAGNGVGRVWIAKD 101
A + I+ IAAGNGVG++ + K+
Sbjct 134 ASQIIIKIAAGNGVGQILVGKE 155
> cpv:cgd2_3260 phosphoglucomutase, tandemly duplicated gene (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=568
Score = 91.3 bits (225), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 40/77 (51%), Positives = 54/77 (70%), Gaps = 0/77 (0%)
Query 21 TPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSDA 80
TP+ QKPGTSGLR+KTR FM+ YL+N++++ F P G T+LVAGDGR+F +A
Sbjct 7 TPYLDQKPGTSGLRKKTRVFMEGTYLANFIESYFQSFPPENFEGATLLVAGDGRFFLPEA 66
Query 81 IKKIVAIAAGNGVGRVW 97
I+ I IAA + V R+W
Sbjct 67 IQIISEIAAAHKVKRIW 83
> cpv:cgd2_3270 phosphoglucomutase, tandemly duplicated gene (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=670
Score = 89.4 bits (220), Expect = 2e-18, Method: Composition-based stats.
Identities = 39/77 (50%), Positives = 54/77 (70%), Gaps = 0/77 (0%)
Query 21 TPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSDA 80
+P+ QKPGTSGLR+KTR FM+ YL+N++++ F P G T+LVAGDGR+F +A
Sbjct 109 SPYLDQKPGTSGLRKKTRVFMEGTYLANFIESYFQSFPPENFEGATLLVAGDGRFFLPEA 168
Query 81 IKKIVAIAAGNGVGRVW 97
I+ I IAA + V R+W
Sbjct 169 IQIISEIAAAHKVKRIW 185
> hsa:5236 PGM1, GSD14; phosphoglucomutase 1 (EC:5.4.2.2); K01835
phosphoglucomutase [EC:5.4.2.2]
Length=580
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 40/84 (47%), Positives = 59/84 (70%), Gaps = 1/84 (1%)
Query 19 ATTPFEGQKPGTSGLRRKTREFMQ-PGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFC 77
AT P+ QKPGTSGLR+KT F + P YL N++Q++F + + +G +++V GDGRYF
Sbjct 26 ATAPYHDQKPGTSGLRKKTYYFEEKPCYLENFIQSIFFSIDLKDRQGSSLVVGGDGRYFN 85
Query 78 SDAIKKIVAIAAGNGVGRVWIAKD 101
AI+ IV +AA NG+GR+ I ++
Sbjct 86 KSAIETIVQMAAANGIGRLVIGQN 109
> cel:R05F9.6 hypothetical protein; K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=568
Score = 81.6 bits (200), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 37/82 (45%), Positives = 53/82 (64%), Gaps = 0/82 (0%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSD 79
T PF GQKPGTSGLR++ EF Q Y N+VQA+ D S+ +G ++V GDGR+ +
Sbjct 10 TKPFAGQKPGTSGLRKRVPEFQQQNYTENFVQAILDAGLGSKKKGSQLVVGGDGRFLSME 69
Query 80 AIKKIVAIAAGNGVGRVWIAKD 101
A I+ IAA NG+ R+ + ++
Sbjct 70 ATNVIIKIAAANGLSRLIVGQN 91
> sce:YMR105C PGM2, GAL5; Pgm2p (EC:5.4.2.2); K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=569
Score = 80.9 bits (198), Expect = 9e-16, Method: Composition-based stats.
Identities = 37/82 (45%), Positives = 56/82 (68%), Gaps = 2/82 (2%)
Query 20 TTPFEGQKPGTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T P+E QKPGTSGLR+KT+ F +P Y N++Q++ + +PE +G T++V GDGRY+
Sbjct 10 TKPYEDQKPGTSGLRKKTKVFKDEPNYTENFIQSIMEAIPEGS-KGATLVVGGDGRYYND 68
Query 79 DAIKKIVAIAAGNGVGRVWIAK 100
+ KI AI A NG+ ++ I +
Sbjct 69 VILHKIAAIGAANGIKKLVIGQ 90
> dre:394000 pgm1, MGC63718, zgc:63718; phosphoglucomutase 1 (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=561
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 57/87 (65%), Gaps = 1/87 (1%)
Query 16 SVRATTPFEGQKPGTSGLRRKTREFMQ-PGYLSNYVQAVFDCLPESEVRGGTMLVAGDGR 74
+V T P+ QKPGTSGLR++ F Q Y N++Q++ + ++ + G ++V GDGR
Sbjct 5 TVIKTKPYTDQKPGTSGLRKRVTVFQQNQHYAENFIQSIISTVDPAQRQEGALVVGGDGR 64
Query 75 YFCSDAIKKIVAIAAGNGVGRVWIAKD 101
+F DAI+ I+ IAA NG+GR+ I +D
Sbjct 65 FFMKDAIQLIIQIAAANGIGRLVIGQD 91
> dre:560297 novel protein similar to vertebrate phosphoglucomutase
5 (PGM5)
Length=567
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 39/83 (46%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Query 20 TTPFEGQKPGTSGLRRKTREF-MQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
TTPF+ Q+PGT+GLR+KT F YL NY+Q+V + + +G TM+V DGRYF
Sbjct 13 TTPFDDQRPGTNGLRKKTAVFESNNNYLQNYIQSVLSSIDLRDRQGCTMVVGSDGRYFSR 72
Query 79 DAIKKIVAIAAGNGVGRVWIAKD 101
A + IV +AA NG+GR+ I +
Sbjct 73 AATEVIVQMAAANGIGRLVIGHN 95
> xla:379864 pgm1, MGC53760, pgm2; phosphoglucomutase 1 (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=562
Score = 77.8 bits (190), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Query 20 TTPFEGQKPGTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T P+ QKPGTSGLR++ F Y N++Q++ C +E + G ++V GDGR++
Sbjct 9 TKPYTDQKPGTSGLRKRVTVFQTNANYAENFIQSIISCTEPAERQDGVLVVGGDGRFYMK 68
Query 79 DAIKKIVAIAAGNGVGRVWIAKD 101
+AI+ I+ IAA NG+GR+ I ++
Sbjct 69 EAIQLIIQIAAANGIGRLVIGQN 91
> sce:YKL127W PGM1; Pgm1p (EC:5.4.2.2); K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=570
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 35/82 (42%), Positives = 52/82 (63%), Gaps = 2/82 (2%)
Query 20 TTPFEGQKPGTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T ++ QKPGTSGLR+KT+ FM +P Y N++QA +P G T++V GDGR++
Sbjct 10 TVAYKDQKPGTSGLRKKTKVFMDEPHYTENFIQATMQSIPNGS-EGTTLVVGGDGRFYND 68
Query 79 DAIKKIVAIAAGNGVGRVWIAK 100
+ KI A+ A NGV ++ I +
Sbjct 69 VIMNKIAAVGAANGVRKLVIGQ 90
> mmu:72157 Pgm2, 2610020G18Rik, AA407108, AI098105, Pgm-2, Pgm1;
phosphoglucomutase 2 (EC:5.4.2.2); K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=562
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Query 20 TTPFEGQKPGTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T + QKPGTSGLR++ + F Y N++Q++ + + + T++V GDGR++ +
Sbjct 9 TQAYPDQKPGTSGLRKRVKVFQSNANYAENFIQSIVSTVEPALRQEATLVVGGDGRFYMT 68
Query 79 DAIKKIVAIAAGNGVGRVWIAKD 101
+AI+ IV IAA NG+GR+ I ++
Sbjct 69 EAIQLIVRIAAANGIGRLVIGQN 91
> hsa:5239 PGM5, PGMRP; phosphoglucomutase 5
Length=567
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/84 (44%), Positives = 53/84 (63%), Gaps = 2/84 (2%)
Query 20 TTPFEGQKP-GTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFC 77
T P+E Q+P G GLRR T F Q YL N++Q+V + + +G TM+V DGRYF
Sbjct 13 TAPYEDQRPAGGGGLRRPTGLFEGQRNYLPNFIQSVLSSIDLRDRQGCTMVVGSDGRYFS 72
Query 78 SDAIKKIVAIAAGNGVGRVWIAKD 101
AI+ +V +AA NG+GR+ I ++
Sbjct 73 RTAIEIVVQMAAANGIGRLIIGQN 96
> mmu:226041 Pgm5, 4833423B07, 9530034F03Rik, D830025G17, aciculin;
phosphoglucomutase 5
Length=567
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/84 (42%), Positives = 52/84 (61%), Gaps = 2/84 (2%)
Query 20 TTPFEGQKP-GTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFC 77
T P+E Q+P G GLRR T F Q YL N++Q+V + + +G TM+V DGRYF
Sbjct 13 TAPYEDQRPTGGGGLRRPTGLFEGQRNYLPNFIQSVLSSIDLRDRQGCTMVVGSDGRYFS 72
Query 78 SDAIKKIVAIAAGNGVGRVWIAKD 101
A + +V +AA NG+GR+ I ++
Sbjct 73 RTATEIVVQMAAANGIGRLIIGQN 96
> hsa:729468 putative PGM5-like protein 1-like
Length=259
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 43/82 (52%), Gaps = 7/82 (8%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSD 79
T P E ++P +GL + R Y N++++ + + +G T V DGRYF
Sbjct 106 TAPCEDRRP--TGLFQGQRS-----YPPNFIRSALSSVDLRDRQGRTTGVGSDGRYFGRT 158
Query 80 AIKKIVAIAAGNGVGRVWIAKD 101
A++ +AA NG+GR+ I ++
Sbjct 159 AVEVGGPMAAANGIGRLIIGQN 180
> hsa:100287382 putative PGM5-like protein 1-like
Length=386
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 34 RRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSDAIKKIVAIAAGNG 92
RR T F Q Y N++++ + + +G T DGR+F A++ + +AA NG
Sbjct 227 RRPTGLFQGQRSYPPNFIRSALSSVGLRDRQGRTTGAGSDGRFFGRTAVEVVGPMAAANG 286
Query 93 VGRVWIAKD 101
+GR+ I ++
Sbjct 287 IGRLIIGQN 295
> xla:398665 ndufa9, MGC64316, ndufs2l; NADH dehydrogenase (ubiquinone)
1 alpha subcomplex, 9, 39kDa; K03953 NADH dehydrogenase
(ubiquinone) 1 alpha subcomplex 9 [EC:1.6.5.3 1.6.99.3]
Length=377
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 42 QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSDAIKKIVAIA 88
QP Y+ + +A+ + + + E G T +AG RY D ++ + A+A
Sbjct 238 QPVYVVDVAKAILNAIHDPESNGKTYALAGPNRYLLHDLVEYVFAVA 284
> mmu:243277 Gpr133, E230012M21Rik; G protein-coupled receptor
133; K08465 G protein-coupled receptor 133
Length=903
Score = 31.6 bits (70), Expect = 0.71, Method: Composition-based stats.
Identities = 16/62 (25%), Positives = 25/62 (40%), Gaps = 0/62 (0%)
Query 7 FSGVPPESASVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGT 66
S PP + P + P + L + + F +PG + Y+Q V LP + T
Sbjct 286 LSSTPPAMPTAHTVIPTDAYHPIITNLTEERKRFQRPGTVLRYLQNVSLRLPNKSLSEET 345
Query 67 ML 68
L
Sbjct 346 AL 347
> mmu:74330 Dnajc14, 5730551F12Rik, DNAJ, DRIP78, HDJ3, LIP6;
DnaJ (Hsp40) homolog, subfamily C, member 14; K09534 DnaJ homolog
subfamily C member 14
Length=703
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query 9 GVPPESASVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLP 58
G+ P++ V F + PGTSG +R T E P L +++ +F P
Sbjct 608 GISPDTHRVPYHISFGSRVPGTSGRQRATPE-SPPADLQDFLSRIFQVPP 656
> mmu:243529 H1fx, Gm461, H1X; H1 histone family, member X; K11275
histone H1/5
Length=188
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 10 VPPESA--SVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTM 67
+PP SA + R T G T RRK R+ QPG Y Q V + + + RGG+
Sbjct 9 LPPTSADGTARKTAKAGGSAAPTQPKRRKNRKKNQPG---KYSQLVVETIRKLGERGGSS 65
Query 68 LV 69
L
Sbjct 66 LA 67
> pfa:PF11_0063 conserved Plasmodium protein
Length=463
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 9/23 (39%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 39 EFMQPGYLSNYVQAVFDCLPESE 61
++ GY SNY+ +F CL E++
Sbjct 149 KYQNKGYCSNYINTIFACLVENK 171
Lambda K H
0.319 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40