bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3112_orf1
Length=89
Score E
Sequences producing significant alignments: (Bits) Value
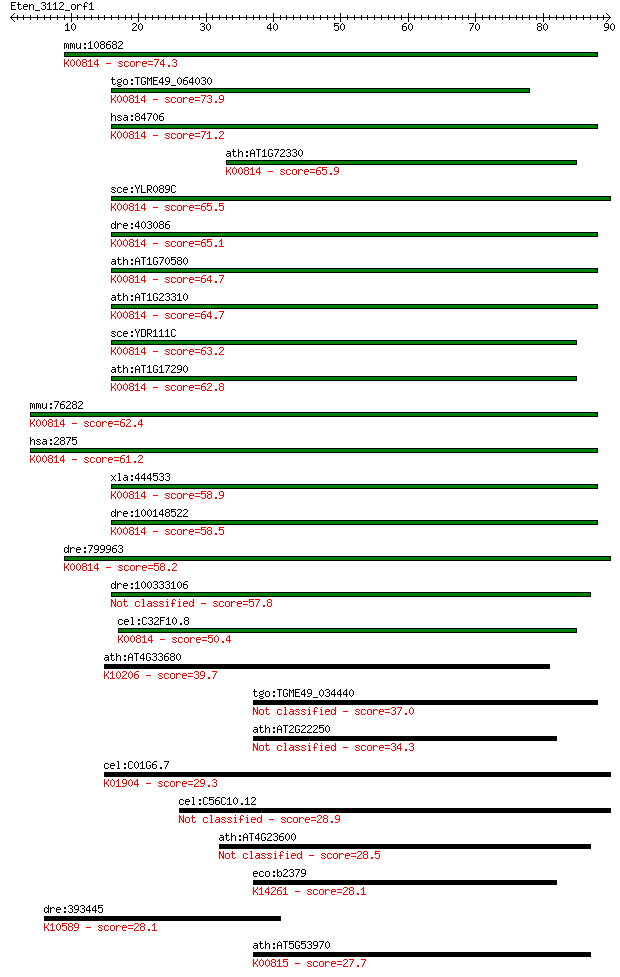
mmu:108682 Gpt2, 4631422C05Rik, ALT2, AU014768, AU041193, C872... 74.3 8e-14
tgo:TGME49_064030 aminotransferase, putative (EC:2.6.1.2); K00... 73.9 1e-13
hsa:84706 GPT2, ALT2; glutamic pyruvate transaminase (alanine ... 71.2 8e-13
ath:AT1G72330 ALAAT2; ALAAT2 (ALANINE AMINOTRANSFERASE 2); ATP... 65.9 3e-11
sce:YLR089C ALT1; Alanine transaminase (glutamic pyruvic trans... 65.5 4e-11
dre:403086 gpt2l, gpt2, im:6791811, wu:fd46f01; glutamic pyruv... 65.1 6e-11
ath:AT1G70580 AOAT2; AOAT2 (ALANINE-2-OXOGLUTARATE AMINOTRANSF... 64.7 7e-11
ath:AT1G23310 GGT1; GGT1 (GLUTAMATE:GLYOXYLATE AMINOTRANSFERAS... 64.7 8e-11
sce:YDR111C ALT2; Alt2p (EC:2.6.1.2); K00814 alanine transamin... 63.2 2e-10
ath:AT1G17290 AlaAT1; AlaAT1 (ALANINE AMINOTRANSFERAS); ATP bi... 62.8 3e-10
mmu:76282 Gpt, 1300007J06Rik, 2310022B03Rik, ALT, Gpt-1, Gpt1;... 62.4 3e-10
hsa:2875 GPT, AAT1, ALT1, GPT1; glutamic-pyruvate transaminase... 61.2 7e-10
xla:444533 gpt2, MGC82097; glutamic pyruvate transaminase (ala... 58.9 4e-09
dre:100148522 Alanine aminotransferase 2-like; K00814 alanine ... 58.5 4e-09
dre:799963 gpt2, MGC165657, im:7150662, sb:cb580, zgc:165657; ... 58.2 7e-09
dre:100333106 glutamic pyruvate transaminase 2-like 57.8 9e-09
cel:C32F10.8 hypothetical protein; K00814 alanine transaminase... 50.4 1e-06
ath:AT4G33680 AGD2; AGD2 (ABERRANT GROWTH AND DEATH 2); L,L-di... 39.7 0.002
tgo:TGME49_034440 aminotransferase, putative (EC:2.6.1.1) 37.0 0.017
ath:AT2G22250 AAT; aminotransferase class I and II family protein 34.3 0.086
cel:C01G6.7 hypothetical protein; K01904 4-coumarate--CoA liga... 29.3 3.4
cel:C56C10.12 hypothetical protein 28.9 4.3
ath:AT4G23600 CORI3; CORI3 (CORONATINE INDUCED 1); cystathioni... 28.5 5.4
eco:b2379 alaC, ECK2375, JW2376, yfdZ; valine-pyruvate aminotr... 28.1 6.5
dre:393445 ube3c, MGC63649, zgc:63649; ubiquitin protein ligas... 28.1 6.6
ath:AT5G53970 aminotransferase, putative (EC:2.6.1.5); K00815 ... 27.7 9.4
> mmu:108682 Gpt2, 4631422C05Rik, ALT2, AU014768, AU041193, C87201;
glutamic pyruvate transaminase (alanine aminotransferase)
2 (EC:2.6.1.2); K00814 alanine transaminase [EC:2.6.1.2]
Length=522
Score = 74.3 bits (181), Expect = 8e-14, Method: Composition-based stats.
Identities = 38/82 (46%), Positives = 54/82 (65%), Gaps = 3/82 (3%)
Query 9 PSAFRVP-FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERDGVPASPD 65
P+ P FPED K+AR L + G S+G+YS SQGV +RE +A + RDGVPA PD
Sbjct 119 PNLLNSPSFPEDAKKRARRILQACGGNSLGSYSASQGVNCIREDVAAFITRRDGVPADPD 178
Query 66 DLFLTDGASAAVARIIEVLYRG 87
+++LT GAS ++ I+++L G
Sbjct 179 NIYLTTGASDGISTILKLLVSG 200
> tgo:TGME49_064030 aminotransferase, putative (EC:2.6.1.2); K00814
alanine transaminase [EC:2.6.1.2]
Length=636
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 33/62 (53%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 16 FPEDVTKKAREYLASVGSVGAYSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGASA 75
F D+ K+R YL ++GSVGAY+HSQG R+ IA W +RDGV PD +FLTDGAS+
Sbjct 249 FALDIVAKSRRYLQAMGSVGAYTHSQGHPLFRKDIAAWLTDRDGVATDPDTIFLTDGASS 308
Query 76 AV 77
+
Sbjct 309 GI 310
> hsa:84706 GPT2, ALT2; glutamic pyruvate transaminase (alanine
aminotransferase) 2 (EC:2.6.1.2); K00814 alanine transaminase
[EC:2.6.1.2]
Length=423
Score = 71.2 bits (173), Expect = 8e-13, Method: Composition-based stats.
Identities = 36/75 (48%), Positives = 51/75 (68%), Gaps = 3/75 (4%)
Query 16 FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERDG-VPASPDDLFLTDG 72
FPED K+AR L + G S+G+YS SQGV +RE +A + RDG VPA PD+++LT G
Sbjct 27 FPEDAKKRARRILQACGGNSLGSYSASQGVNCIREDVAAYITRRDGGVPADPDNIYLTTG 86
Query 73 ASAAVARIIEVLYRG 87
AS ++ I+++L G
Sbjct 87 ASDGISTILKILVSG 101
> ath:AT1G72330 ALAAT2; ALAAT2 (ALANINE AMINOTRANSFERASE 2); ATP
binding / L-alanine:2-oxoglutarate aminotransferase; K00814
alanine transaminase [EC:2.6.1.2]
Length=431
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 29/52 (55%), Positives = 40/52 (76%), Gaps = 0/52 (0%)
Query 33 SVGAYSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGASAAVARIIEVL 84
+ GAYSHSQG++ LR++IA RDG PA P+D+FLTDGAS AV ++++L
Sbjct 166 ATGAYSHSQGIKGLRDVIAAGIEARDGFPADPNDIFLTDGASPAVHMMMQLL 217
> sce:YLR089C ALT1; Alanine transaminase (glutamic pyruvic transaminase);
involved in alanine biosynthetic and catabolic processes;
the authentic, non-tagged protein is detected in highly
purified mitochondria in high-throughput studies (EC:2.6.1.2);
K00814 alanine transaminase [EC:2.6.1.2]
Length=592
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/76 (46%), Positives = 50/76 (65%), Gaps = 2/76 (2%)
Query 16 FPEDVTKKAREYLASVG-SVGAYSHSQGVRELREIIATWFGERD-GVPASPDDLFLTDGA 73
F D K+A+ + +G SVGAYS SQGV +R+ +A + +RD G + P+D+FLT GA
Sbjct 199 FKLDAIKRAKSLMEDIGGSVGAYSSSQGVEGIRKSVAEFITKRDEGEISYPEDIFLTAGA 258
Query 74 SAAVARIIEVLYRGPE 89
SAAV ++ + RGPE
Sbjct 259 SAAVNYLLSIFCRGPE 274
> dre:403086 gpt2l, gpt2, im:6791811, wu:fd46f01; glutamic pyruvate
transaminase (alanine aminotransferase) 2, like (EC:2.6.1.2);
K00814 alanine transaminase [EC:2.6.1.2]
Length=493
Score = 65.1 bits (157), Expect = 6e-11, Method: Composition-based stats.
Identities = 32/75 (42%), Positives = 49/75 (65%), Gaps = 3/75 (4%)
Query 16 FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERDG-VPASPDDLFLTDG 72
FPED +AR L S G S+GAY+ SQG+ +R+ +A + RDG +P+ PD+++LT G
Sbjct 97 FPEDAKNRARRILQSCGGNSIGAYTTSQGIDCVRQDVAKYIERRDGGIPSDPDNIYLTTG 156
Query 73 ASAAVARIIEVLYRG 87
AS + I+++L G
Sbjct 157 ASDGIVTILKLLTAG 171
> ath:AT1G70580 AOAT2; AOAT2 (ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE
2); L-alanine:2-oxoglutarate aminotransferase/ glycine:2-oxoglutarate
aminotransferase (EC:2.6.1.2); K00814 alanine
transaminase [EC:2.6.1.2]
Length=481
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 31/73 (42%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Query 16 FPEDVTKKAREYLA-SVGSVGAYSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGAS 74
FP D +A+ YL+ + G +GAYS S+G+ +R+ +A + RDG P+ P+ +FLTDGAS
Sbjct 81 FPADAIARAKHYLSLTSGGLGAYSDSRGLPGVRKEVAEFIERRDGYPSDPELIFLTDGAS 140
Query 75 AAVARIIEVLYRG 87
V +I+ + RG
Sbjct 141 KGVMQILNCVIRG 153
> ath:AT1G23310 GGT1; GGT1 (GLUTAMATE:GLYOXYLATE AMINOTRANSFERASE);
L-alanine:2-oxoglutarate aminotransferase/ glycine:2-oxoglutarate
aminotransferase (EC:2.6.1.2); K00814 alanine transaminase
[EC:2.6.1.2]
Length=441
Score = 64.7 bits (156), Expect = 8e-11, Method: Composition-based stats.
Identities = 31/73 (42%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Query 16 FPEDVTKKAREYLA-SVGSVGAYSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGAS 74
FP D +A+ YL+ + G +GAYS S+G+ +R+ +A + RDG P+ P+ +FLTDGAS
Sbjct 81 FPADAIARAKHYLSLTSGGLGAYSDSRGLPGVRKEVAEFIQRRDGYPSDPELIFLTDGAS 140
Query 75 AAVARIIEVLYRG 87
V +I+ + RG
Sbjct 141 KGVMQILNCVIRG 153
> sce:YDR111C ALT2; Alt2p (EC:2.6.1.2); K00814 alanine transaminase
[EC:2.6.1.2]
Length=507
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 31/71 (43%), Positives = 47/71 (66%), Gaps = 2/71 (2%)
Query 16 FPEDVTKKAREYLASVG-SVGAYSHSQGVRELREIIATWFGERD-GVPASPDDLFLTDGA 73
F D ++A L +G S+GAYSHSQGV +R+ +A + RD G PA+P+D++LT GA
Sbjct 114 FSRDALERAERLLNDIGGSIGAYSHSQGVPGIRQTVADFITRRDGGEPATPEDIYLTTGA 173
Query 74 SAAVARIIEVL 84
S+A ++ +L
Sbjct 174 SSAATSLLSLL 184
> ath:AT1G17290 AlaAT1; AlaAT1 (ALANINE AMINOTRANSFERAS); ATP
binding / L-alanine:2-oxoglutarate aminotransferase (EC:2.6.1.2);
K00814 alanine transaminase [EC:2.6.1.2]
Length=543
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 31/71 (43%), Positives = 46/71 (64%), Gaps = 2/71 (2%)
Query 16 FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGA 73
F D ++A + L + + GAYSHSQG++ LR+ IA RDG PA P+D+F+TDGA
Sbjct 150 FSSDSIERAWKILDQIPGRATGAYSHSQGIKGLRDAIADGIEARDGFPADPNDIFMTDGA 209
Query 74 SAAVARIIEVL 84
S V ++++L
Sbjct 210 SPGVHMMMQLL 220
> mmu:76282 Gpt, 1300007J06Rik, 2310022B03Rik, ALT, Gpt-1, Gpt1;
glutamic pyruvic transaminase, soluble (EC:2.6.1.2); K00814
alanine transaminase [EC:2.6.1.2]
Length=496
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 35/88 (39%), Positives = 55/88 (62%), Gaps = 4/88 (4%)
Query 4 ALDFCPSAFRVP-FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERDG- 59
AL P+ P FPED ++A L + G S+GAYS S G++ +RE +A + RDG
Sbjct 87 ALCVYPNLLSSPDFPEDAKRRAERILQACGGHSLGAYSISSGIQPIREDVAQYIERRDGG 146
Query 60 VPASPDDLFLTDGASAAVARIIEVLYRG 87
+PA P+++FL+ GAS A+ ++++L G
Sbjct 147 IPADPNNIFLSTGASDAIVTMLKLLVAG 174
> hsa:2875 GPT, AAT1, ALT1, GPT1; glutamic-pyruvate transaminase
(alanine aminotransferase) (EC:2.6.1.2); K00814 alanine transaminase
[EC:2.6.1.2]
Length=496
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 35/88 (39%), Positives = 54/88 (61%), Gaps = 4/88 (4%)
Query 4 ALDFCPSAFRVP-FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERDG- 59
AL P P FP+D K+A L + G S+GAYS S G++ +RE +A + RDG
Sbjct 87 ALCVNPDLLSSPNFPDDAKKRAERILQACGGHSLGAYSVSSGIQLIREDVARYIERRDGG 146
Query 60 VPASPDDLFLTDGASAAVARIIEVLYRG 87
+PA P+++FL+ GAS A+ ++++L G
Sbjct 147 IPADPNNVFLSTGASDAIVTVLKLLVAG 174
> xla:444533 gpt2, MGC82097; glutamic pyruvate transaminase (alanine
aminotransferase) 2 (EC:2.6.1.2); K00814 alanine transaminase
[EC:2.6.1.2]
Length=540
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 29/75 (38%), Positives = 50/75 (66%), Gaps = 3/75 (4%)
Query 16 FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERD-GVPASPDDLFLTDG 72
FPEDV +KA L + G S+GAYS SQG+ +R+ +A + RD G+ + P++++L+ G
Sbjct 144 FPEDVKQKAARILQACGGHSIGAYSASQGIEVIRQDVAKYIERRDGGILSDPNNIYLSTG 203
Query 73 ASAAVARIIEVLYRG 87
AS ++ ++++L G
Sbjct 204 ASDSIVTMLKLLVSG 218
> dre:100148522 Alanine aminotransferase 2-like; K00814 alanine
transaminase [EC:2.6.1.2]
Length=488
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 28/75 (37%), Positives = 48/75 (64%), Gaps = 3/75 (4%)
Query 16 FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERD-GVPASPDDLFLTDG 72
FPED +A+ L + G S+GAYS SQG+ +R+ +A + RD G+ PD+++L+ G
Sbjct 92 FPEDAKSRAQRILKACGGGSLGAYSTSQGIEMIRQDVARYIERRDGGIACDPDNIYLSTG 151
Query 73 ASAAVARIIEVLYRG 87
AS A+ +++++ G
Sbjct 152 ASDAIVTMLKLMVSG 166
> dre:799963 gpt2, MGC165657, im:7150662, sb:cb580, zgc:165657;
glutamic pyruvate transaminase (alanine aminotransferase)
2 (EC:2.6.1.2); K00814 alanine transaminase [EC:2.6.1.2]
Length=484
Score = 58.2 bits (139), Expect = 7e-09, Method: Composition-based stats.
Identities = 34/85 (40%), Positives = 50/85 (58%), Gaps = 4/85 (4%)
Query 9 PSAFRVP-FPEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERD-GVPASP 64
P P FPED +AR L G S+G+YS S GV +R+ IA + +RD GVP++
Sbjct 77 PELMESPSFPEDAKWRARRILQGCGGHSLGSYSASAGVEYIRKDIAAYIEQRDEGVPSNW 136
Query 65 DDLFLTDGASAAVARIIEVLYRGPE 89
+D++LT GAS + I+ +L G +
Sbjct 137 EDIYLTTGASDGIMTILRLLVSGKD 161
> dre:100333106 glutamic pyruvate transaminase 2-like
Length=470
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 47/74 (63%), Gaps = 3/74 (4%)
Query 16 FPEDVTKKAREYLASV--GSVGAYSHSQGVRELREIIATWFGERD-GVPASPDDLFLTDG 72
P DV ++A + L GSVG+YS S G+ ++RE + + RD GV +SPD++F++ G
Sbjct 74 LPSDVRQRAEDLLRECAGGSVGSYSESSGMAKVRECVCEFISRRDGGVSSSPDNIFISTG 133
Query 73 ASAAVARIIEVLYR 86
+ A+ +++ VL R
Sbjct 134 SQTALMKLLTVLIR 147
> cel:C32F10.8 hypothetical protein; K00814 alanine transaminase
[EC:2.6.1.2]
Length=504
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 43/71 (60%), Gaps = 3/71 (4%)
Query 17 PEDVTKKAREYLASVG--SVGAYSHSQGVRELREIIATWFGERD-GVPASPDDLFLTDGA 73
P DV + A +L S G S GAYS S GV +R+ +A + RD G+P + +D+ L+ GA
Sbjct 110 PSDVIEHANAFLGSCGGKSAGAYSQSTGVEIVRKHVAEYIKRRDGGIPCNSEDVCLSGGA 169
Query 74 SAAVARIIEVL 84
S ++ ++++
Sbjct 170 SESIRNVLKLF 180
> ath:AT4G33680 AGD2; AGD2 (ABERRANT GROWTH AND DEATH 2); L,L-diaminopimelate
aminotransferase/ transaminase; K10206 LL-diaminopimelate
aminotransferase [EC:2.6.1.83]
Length=461
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 40/71 (56%), Gaps = 9/71 (12%)
Query 15 PFPEDVT----KKAREYLASVGSVGAYSHSQGVRELREIIA-TWFGERDGVPASPDDLFL 69
P PE +T KKA E L+++ Y QG + LR IA T++G G+ DD+F+
Sbjct 104 PIPEVITSAMAKKAHE-LSTIEGYSGYGAEQGAKPLRAAIAKTFYG---GLGIGDDDVFV 159
Query 70 TDGASAAVARI 80
+DGA ++R+
Sbjct 160 SDGAKCDISRL 170
> tgo:TGME49_034440 aminotransferase, putative (EC:2.6.1.1)
Length=411
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 37 YSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGASAAVARIIEVLYRG 87
Y +G E RE +A++ E G P S D+L +G+S A+ I +V +G
Sbjct 69 YGVPEGFLEFRESLASFLSEEYGFPVSSDELMAVNGSSGALDLICKVYTQG 119
> ath:AT2G22250 AAT; aminotransferase class I and II family protein
Length=475
Score = 34.3 bits (77), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 37 YSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGASAAVARII 81
Y+ + G+ ELRE I E +G+ +PD + +++GA ++ + +
Sbjct 132 YTLNAGITELREAICRKLKEENGLSYAPDQILVSNGAKQSLLQAV 176
> cel:C01G6.7 hypothetical protein; K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=540
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 26/83 (31%), Positives = 36/83 (43%), Gaps = 14/83 (16%)
Query 15 PFPEDVTKKAREYLASVGSVGAYSHSQGVRELREIIATWF---GERDG-----VPASPDD 66
PFPE +KK ++ L +V V Y G+ EL AT G DG VP +
Sbjct 313 PFPESASKKLKQLLPNVNIVQGY----GMTEL--TFATHLQSPGSPDGSVGRLVPGTSMK 366
Query 67 LFLTDGASAAVARIIEVLYRGPE 89
+ DG I E+ +GP+
Sbjct 367 VKKEDGTLCGPHEIGELWIKGPQ 389
> cel:C56C10.12 hypothetical protein
Length=1599
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 10/65 (15%)
Query 26 EYLA-SVGSVGAYSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGASAAVARIIEVL 84
E+L S+ S+G+YS+S + L+E+I F + D + DDL+ G + I ++L
Sbjct 712 EFLVNSLASIGSYSNSDVSQLLKELIDVCFCDED----TRDDLYKCGGEA-----IGQIL 762
Query 85 YRGPE 89
+ PE
Sbjct 763 IKRPE 767
> ath:AT4G23600 CORI3; CORI3 (CORONATINE INDUCED 1); cystathionine
beta-lyase/ transaminase
Length=380
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 31/57 (54%), Gaps = 4/57 (7%)
Query 32 GSVGAYSHSQGVRELREIIATWFGERDGVPA--SPDDLFLTDGASAAVARIIEVLYR 86
GS AY+ S G+ + +A + + G+P + DD+F+T G A+ +++L +
Sbjct 68 GSGNAYAPSLGLAAAKSAVAEYLNQ--GLPKKLTADDVFMTLGCKQAIELAVDILAK 122
> eco:b2379 alaC, ECK2375, JW2376, yfdZ; valine-pyruvate aminotransferase
3; K14261 alanine-synthesizing transaminase [EC:2.6.1.-]
Length=412
Score = 28.1 bits (61), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 37 YSHSQGVRELREIIATWFGERDGVPASPD-DLFLTDGASAAVARII 81
YS S+G+ LR I+ W+ +R V P+ + +T G+ +A ++
Sbjct 70 YSTSRGIPRLRRAISRWYQDRYDVEIDPESEAIVTIGSKEGLAHLM 115
> dre:393445 ube3c, MGC63649, zgc:63649; ubiquitin protein ligase
E3C; K10589 ubiquitin-protein ligase E3 C [EC:6.3.2.19]
Length=994
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 6 DFCPSAFRVPFPEDVTKKAREYLASVGSVGAYSHS 40
D C ++ +PE T+ EY+A+ SVG ++S
Sbjct 549 DACLGIIKLAYPETKTEHKEEYVAAFRSVGVKTNS 583
> ath:AT5G53970 aminotransferase, putative (EC:2.6.1.5); K00815
tyrosine aminotransferase [EC:2.6.1.5]
Length=414
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 37 YSHSQGVRELREIIATWFGERDGVPASPDDLFLTDGASAAVARIIEVLYR 86
YS + G+ + R IA + S DD+F+T G + A+ + +L R
Sbjct 71 YSPTVGLPQARRAIAEYLSRDLPYKLSQDDVFITSGCTQAIDVALSMLAR 120
Lambda K H
0.318 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017039696
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40