bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3108_orf1
Length=437
Score E
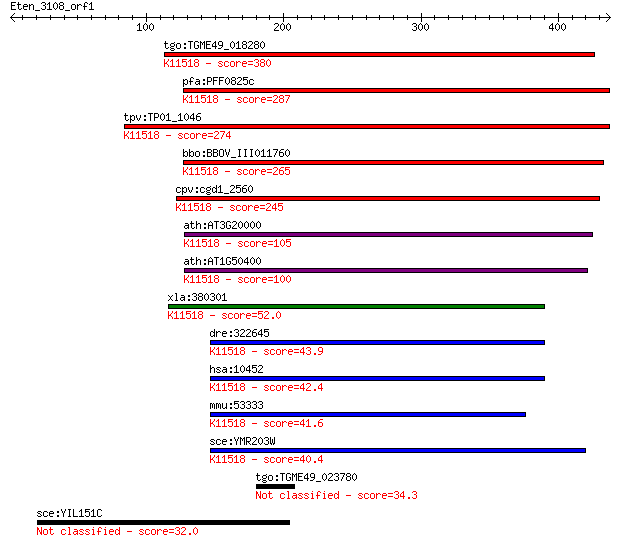
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_018280 eukaryotic porin domain-containing protein ;... 380 6e-105
pfa:PFF0825c mitochondrial import receptor subunit tom40; K115... 287 6e-77
tpv:TP01_1046 mitochondrial import receptor subunit Tom40; K11... 274 5e-73
bbo:BBOV_III011760 17.m08003; hypothetical protein; K11518 mit... 265 2e-70
cpv:cgd1_2560 Tom40p like translocase ; K11518 mitochondrial i... 245 2e-64
ath:AT3G20000 TOM40; TOM40; P-P-bond-hydrolysis-driven protein... 105 4e-22
ath:AT1G50400 porin family protein; K11518 mitochondrial impor... 100 2e-20
xla:380301 tomm40, MGC53384; translocase of outer mitochondria... 52.0 4e-06
dre:322645 tomm40, wu:fb68h07, zgc:64168; translocase of outer... 43.9 0.001
hsa:10452 TOMM40, C19orf1, D19S1177E, PER-EC1, PEREC1, TOM40; ... 42.4 0.003
mmu:53333 Tomm40, AW539759, MGC118268, Mom35, Tom40; transloca... 41.6 0.007
sce:YMR203W TOM40, ISP42, MOM38; Component of the TOM (translo... 40.4 0.016
tgo:TGME49_023780 hypothetical protein 34.3 0.91
sce:YIL151C Putative protein of unknown function, predicted to... 32.0 5.2
> tgo:TGME49_018280 eukaryotic porin domain-containing protein
; K11518 mitochondrial import receptor subunit TOM40
Length=466
Score = 380 bits (976), Expect = 6e-105, Method: Compositional matrix adjust.
Identities = 172/318 (54%), Positives = 227/318 (71%), Gaps = 8/318 (2%)
Query 113 NEQTPEPPKKPQPTQ---PIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFC 169
N P K+ T+ P+P+E FSREWM ++GQD FDGFRLE KQVNK L H+F
Sbjct 140 NRLIPSESKELDKTRAPIPVPYEQFSREWMAVAGQDNFDGFRLEATKQVNKVLTANHTFM 199
Query 170 LGTQLRESGYSYQFGPTVAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLC 229
LGTQ +E G SY FGPT+ I P + +Q E P +FF MA++ +DG LQ R IK +
Sbjct 200 LGTQSKEGGCSYSFGPTLVIGEPDEAAQQEGQMP---NFFGMARMNSDGFLQARFIKAIS 256
Query 230 QSADLKLNFNSSLKDEQRN--FYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRL 287
++ D+K N NSS+ ++ ++ YE+S D G DWA++LK+AWQ WIL+G+FSQV+TP+L
Sbjct 257 KTFDIKFNSNSSISEDAKDKSMYEVSFDKMGSDWAANLKLAWQGTWILNGLFSQVITPKL 316
Query 288 QAGCEITYVGANGASMLAVGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYAR 347
Q G E+T+V A G SM +VG RY ++ + V+CQ+ PDF SP G ++ K QY R
Sbjct 317 QLGGELTWVAATGISMGSVGARYGFNENNTVTCQIGVGPDFSSPMGFANDVYSTKAQYVR 376
Query 348 KVTDRLSMASEFEYSHPETESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVSG 407
KVTDRLSM +E E++HP+ SA+RVGW+YLFRQARVQGL+D+ GR+S+FAQDYNGFG+SG
Sbjct 377 KVTDRLSMGTELEFTHPDMSSAMRVGWQYLFRQARVQGLVDTAGRVSMFAQDYNGFGLSG 436
Query 408 LIDYWRGTYKFGFLMHVV 425
+IDYW G YKFGF M+VV
Sbjct 437 MIDYWHGDYKFGFQMNVV 454
> pfa:PFF0825c mitochondrial import receptor subunit tom40; K11518
mitochondrial import receptor subunit TOM40
Length=377
Score = 287 bits (734), Expect = 6e-77, Method: Compositional matrix adjust.
Identities = 137/310 (44%), Positives = 195/310 (62%), Gaps = 19/310 (6%)
Query 127 QPIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSYQFGPT 186
+ FE+ ++E+ I+ QD FDGFR E K +NK+LQ+ H+ LGT LRE GY YQFG
Sbjct 84 NALLFENLNKEYKFITTQDNFDGFRFEVDKNINKFLQSTHTLFLGTTLREVGYLYQFGA- 142
Query 187 VAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQ 246
N ++++ DGS+ GR K + + D KLNFN+ K++
Sbjct 143 -------------NFTNLDNSLLMISRINIDGSVNGRFCKKINNNIDCKLNFNTYAKNDT 189
Query 247 RNFYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVGANGASMLAV 306
RN YE+SL+ + + K WQ WI + ++Q+LT +LQAG ++TY+ +N AS+ +
Sbjct 190 RNMYEMSLEVNKPLYTYNFKSIWQGAWIFNTSYTQLLTKKLQAGVDLTYIASNCASIGSF 249
Query 307 GGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPET 366
G RYN K +V++ Q+ +QP+FKSP + TH K+QYA+K++DRLS+ +E E +
Sbjct 250 GLRYN-HKNNVLTMQIVRQPNFKSPEFMLNQTHLYKIQYAKKISDRLSLGTELEITPQTK 308
Query 367 ESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVSGLIDYWRGTYKFGFLMHVVP 426
ESA+R+GW+Y FR A+VQG ID+ G+ISVF QDY+GFGVSG IDY YKFGF+MH+ P
Sbjct 309 ESAMRLGWDYSFRHAKVQGSIDTSGKISVFTQDYSGFGVSGYIDYLNNDYKFGFMMHISP 368
Query 427 PPEASPSQPQ 436
E QPQ
Sbjct 369 SQE----QPQ 374
> tpv:TP01_1046 mitochondrial import receptor subunit Tom40; K11518
mitochondrial import receptor subunit TOM40
Length=394
Score = 274 bits (701), Expect = 5e-73, Method: Compositional matrix adjust.
Identities = 147/353 (41%), Positives = 207/353 (58%), Gaps = 26/353 (7%)
Query 84 SKESQSASSDAPAAVDKREKARLDTLARENEQTPEPPKKPQPTQPIPFEHFSREWMNISG 143
S+ QS S A + +K+E ++E++Q P+P P + +E+ +RE+ N+
Sbjct 66 SERFQSLLSVAHCSEEKQE-------SKEDQQHPQPS----PFDLLVYENLAREYKNVIT 114
Query 144 QDTFDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSYQFGPTVAISPPPDPSQGENAQP 203
QD +DGFRLE +Q+ K LQ HS LGT L++ GY YQ G A G+
Sbjct 115 QDNYDGFRLEADRQITKNLQASHSLYLGTLLKDVGYIYQIGANYA------SDDGKK--- 165
Query 204 PPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNTGRDWAS 263
F M K+G DG++ + L +LK NS LK + ++ +E+ D W +
Sbjct 166 -----FLMFKVGLDGNIALKAFAKLGNRLELKAVTNSRLKIDNQSTFEVGADYLSDYWTA 220
Query 264 SLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVGANGASMLAVGGRYNLDKQSVVSCQLS 323
+LK WQ +I + ++ + P G E+TY+ ANGAS+ ++ RY + ++ +CQL+
Sbjct 221 TLKCCWQGTFIWNICYTHQILPCFTVGSEVTYIDANGASIGSLSARY-VRGDNIFTCQLT 279
Query 324 QQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPETESALRVGWEYLFRQARV 383
+QPDFK K + +VQY RKV DRLS+A+E E S ESALRVGWEYLFR ARV
Sbjct 280 RQPDFKHMDFSKKEINCARVQYTRKVNDRLSLATELELSPSIKESALRVGWEYLFRHARV 339
Query 384 QGLIDSCGRISVFAQDYNGFGVSGLIDYWRGTYKFGFLMHVVPPPEASPSQPQ 436
QG IDSCGRI++ QDYNGFGVSG IDYW Y+FGF+MH++P PE + PQ
Sbjct 340 QGNIDSCGRIAMQTQDYNGFGVSGCIDYWNNIYRFGFMMHLLPQPEQNQENPQ 392
> bbo:BBOV_III011760 17.m08003; hypothetical protein; K11518 mitochondrial
import receptor subunit TOM40
Length=402
Score = 265 bits (677), Expect = 2e-70, Method: Compositional matrix adjust.
Identities = 138/307 (44%), Positives = 188/307 (61%), Gaps = 17/307 (5%)
Query 127 QPIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSYQFGPT 186
+ + FE+ +RE+ N+ QD +DGFRLE +Q+ LQ HS LGT + E+GY YQ G
Sbjct 106 ELLVFENLTREFKNVITQDNYDGFRLEADRQLTNNLQLSHSLYLGTVMSETGYLYQIGTN 165
Query 187 VAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQ 246
A S + AM K+G DG L GR+ + K++ NS L+ +
Sbjct 166 YASSDG--------------NCLAMGKIGLDGMLTGRVFAKYAD-CEFKVSGNSFLRYDT 210
Query 247 RNFYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVGANGASMLAV 306
RN YE +D G+ W +SLK AWQ WIL+ ++ L P L G E+TY+ ANG S+ A+
Sbjct 211 RNAYEAGVDYFGKGWTASLKGAWQGTWILNAAYTHQLLPCLTLGSELTYIVANGTSIGAI 270
Query 307 GGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPET 366
RY + V + Q S+QP+FK + T TC++QY R+V +RLS+ +E E +
Sbjct 271 AARY-VRGDHVFTGQWSKQPNFKGMDYNLQDTDTCRLQYVRRVNERLSLCTELEVTPATK 329
Query 367 ESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVSGLIDYWRGTYKFGFLMHVVP 426
ESALRVGW+YLFR ARVQG ID+ GRIS+ AQDY+GFG+SG IDYW Y+FGF+MH++P
Sbjct 330 ESALRVGWDYLFRHARVQGNIDTAGRISMQAQDYSGFGISGCIDYWSNIYRFGFMMHLLP 389
Query 427 P-PEASP 432
P PE P
Sbjct 390 PVPEQKP 396
> cpv:cgd1_2560 Tom40p like translocase ; K11518 mitochondrial
import receptor subunit TOM40
Length=309
Score = 245 bits (626), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 122/308 (39%), Positives = 189/308 (61%), Gaps = 16/308 (5%)
Query 122 KPQPTQPIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSY 181
+ + + +E+FS E +I +++FDGFR E AK V LQT +S LGT GYSY
Sbjct 12 NERTNKSLQYENFSLESQSIFQRESFDGFRAEIAKSVTSNLQTSYSLFLGTS-NIKGYSY 70
Query 182 QFGPTVAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSS 241
Q GP+ Q + +A++ +G++ GR + + + +++ NSS
Sbjct 71 QLGPSY--------------QSSSKNTLILARVNDEGTVSGRFSRCFGNNIEGRISINSS 116
Query 242 LKDEQRNFYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVGANGA 301
L DE +N E+S+D G + + SLKVA+Q ++L+G+FSQ++T +LQ G E+T++ AN
Sbjct 117 LSDENKNMSEVSIDYNGEESSYSLKVAYQGIFLLNGLFSQLITNKLQLGGELTWIAANNT 176
Query 302 SMLAVGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEY 361
S++++G RY +++ Q+++QPDF SP + H+ + + RK++DRLS+ASE E
Sbjct 177 SIMSLGSRY-CHGKNIFFNQITRQPDFTSPGRIFANIHSLRSSFYRKLSDRLSLASELEV 235
Query 362 SHPETESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVSGLIDYWRGTYKFGFL 421
S P ES LR G+EYLF+ AR+QG+ID+CG+IS+ D GFG+S IDY R YKFGF+
Sbjct 236 SIPNFESTLRFGYEYLFKTARIQGMIDTCGKISLQCLDNKGFGISAAIDYLRNDYKFGFM 295
Query 422 MHVVPPPE 429
M P +
Sbjct 296 MQFFPNEK 303
> ath:AT3G20000 TOM40; TOM40; P-P-bond-hydrolysis-driven protein
transmembrane transporter/ voltage-gated anion channel; K11518
mitochondrial import receptor subunit TOM40
Length=309
Score = 105 bits (262), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 82/311 (26%), Positives = 136/311 (43%), Gaps = 47/311 (15%)
Query 128 PIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLG----------TQLRES 177
P+P+E RE + D F+G R + + +N+ HS +G T ++
Sbjct 31 PVPYEELHREALMSLKSDNFEGLRFDFTRALNQKFSLSHSVMMGPTEVPAQSPETTIKIP 90
Query 178 GYSYQFGPTVAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLN 237
Y+FG A P + ++ TDG L RL L +K N
Sbjct 91 TAHYEFG----------------ANYYDPKLLLIGRVMTDGRLNARLKADLTDKLVVKAN 134
Query 238 FNSSLKDEQRNFYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVG 297
+ +E + + D G D+ + L++ QS I + Q +T L G EI + G
Sbjct 135 ALIT-NEEHMSQAMFNFDYMGSDYRAQLQLG-QSALI-GATYIQSVTNHLSLGGEIFWAG 191
Query 298 ANGASMLAVGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMAS 357
S + RY DK V S Q++ T + Y +K++D++S+A+
Sbjct 192 VPRKSGIGYAARYETDKM-VASGQVAS-------------TGAVVMNYVQKISDKVSLAT 237
Query 358 EFEYSHPETESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVSGL----IDYWR 413
+F Y++ + VG++Y+ RQARV+G IDS G S ++ G++ L +D+ +
Sbjct 238 DFMYNYFSRDVTASVGYDYMLRQARVRGKIDSNGVASALLEERLSMGLNFLLSAELDHKK 297
Query 414 GTYKFGFLMHV 424
YKFGF + V
Sbjct 298 KDYKFGFGLTV 308
> ath:AT1G50400 porin family protein; K11518 mitochondrial import
receptor subunit TOM40
Length=310
Score = 100 bits (248), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 77/299 (25%), Positives = 141/299 (47%), Gaps = 30/299 (10%)
Query 128 PIPFEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLG-TQLRESGYSYQFGPT 186
P+ +E +RE + F+GFRL+ K +N+ HS +G T++ P+
Sbjct 31 PVLYEELNREATMALKPELFEGFRLDYNKSLNQKFFLSHSILMGPTEVPNPT------PS 84
Query 187 VAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQ 246
I P + A P + + ++ TDG L R L + +K N + L DE+
Sbjct 85 SEIIKIPTANYDFGAGFIDPKLYLIGRITTDGRLNARAKFDLTDNFSVKAN--ALLTDEE 142
Query 247 -RNFYEISLDNTGRDWASSLKVAWQSCWILDGMFSQVLTPRLQAGCEITYVGANGASMLA 305
++ + +D G D+ + L++ S + + + Q +TP L G E ++G S +
Sbjct 143 DKSQGHLVIDYKGSDYRTQLQLGNNSVYAAN--YIQHVTPHLSLGGEAFWLGQQLMSGVG 200
Query 306 VGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPE 365
RY DK +V S Q++ T + Y KV+++LS A++F Y++
Sbjct 201 YAARYETDK-TVASGQIAS-------------TGVAVMNYVHKVSEKLSFATDFIYNYLS 246
Query 366 TESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFG----VSGLIDYWRGTYKFGF 420
+ VG++ + RQ+R++G +DS G ++ + ++ G +S +D+ + YKFGF
Sbjct 247 RDVTASVGYDLITRQSRLRGKVDSNGVVAAYLEEQLPIGLRFLLSAEVDHVKKDYKFGF 305
> xla:380301 tomm40, MGC53384; translocase of outer mitochondrial
membrane 40 homolog; K11518 mitochondrial import receptor
subunit TOM40
Length=336
Score = 52.0 bits (123), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 71/288 (24%), Positives = 119/288 (41%), Gaps = 45/288 (15%)
Query 116 TPEPPKKPQPTQPIP----FEHFSREWMNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLG 171
TP P KK + +P FE R+ + +G +L K ++ Y Q H+ L
Sbjct 39 TPTPDKKEPQEERLPNPGTFEECHRKCKELF-PIQMEGVKLIVNKGLSNYFQVNHTISLS 97
Query 172 TQLRESGYSYQFGPTVAISPPPDPSQGENAQPPPPHFFAMAKLGTDGSLQGRLIKTLCQS 231
T + ES +Y FG T + P++ A P + GSL G++I + +
Sbjct 98 T-IGES--NYHFGATYVGTKQLGPAE---AFP-----VLVGDFDNSGSLNGQIIHQVTNN 146
Query 232 ADLKLNFNSSLKDEQRNFYEISLDNT--GRDWASSLKVA----WQSCWILDGMFSQVLTP 285
K+ +L+ +Q F LD G D+ +S+ + IL + Q +TP
Sbjct 147 IRSKI----ALQTQQSKFVNWQLDTEYRGEDYTASVTLGNPDILVGSGILVAHYLQSITP 202
Query 286 RLQAGCEITYVGANG--ASMLAVGGRYNLDKQSVVSCQLSQQPDFKSPAGLTKLTHTCKV 343
L G E+ Y G +++++ GRY P + A LT
Sbjct 203 SLALGGELVYHRRPGEEGTVMSLAGRYT-------------APSWT--ATLTLGQAGAHA 247
Query 344 QYARKVTDRLSMASEFEYSHPETESALRVGWEYLFRQARV--QGLIDS 389
Y K D+L + EFE S ++++ +G++ +A + +G IDS
Sbjct 248 TYYHKANDQLQLGVEFEASARMQDTSVSLGYQLDLPKANLLFKGSIDS 295
> dre:322645 tomm40, wu:fb68h07, zgc:64168; translocase of outer
mitochondrial membrane 40 homolog (yeast); K11518 mitochondrial
import receptor subunit TOM40
Length=344
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 59/254 (23%), Positives = 99/254 (38%), Gaps = 42/254 (16%)
Query 147 FDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSYQFGPTVAISPPPDPSQGENAQPPPP 206
+G RL K ++ + Q H+ L T L +SGY +FG T G P
Sbjct 81 MEGVRLVVNKGLSNHFQVSHTITLST-LGDSGY--RFGSTYV---------GSKQTGPAE 128
Query 207 HFFAMA-KLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNT--GRDWAS 263
F M + GSL ++I L K+ +++ +Q F D G D+ +
Sbjct 129 SFPVMVGDMDNTGSLNAQVIHQLTNRVRSKI----AIQTQQHKFVNWQCDGEYRGDDFTA 184
Query 264 SLKVAWQSCWILDGM----FSQVLTPRLQAGCEITYVGANG--ASMLAVGGRYNLDKQSV 317
++ + + G+ + Q L+P L G E+ Y G ++ ++ GRY
Sbjct 185 AVTLGNPDVLVGSGILVAHYLQSLSPSLVLGGELVYHKRPGEEGTVTSLVGRYTGSNYV- 243
Query 318 VSCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPETESALRVGWEYL 377
A LT Y K D+L + EFE S E+++ G++
Sbjct 244 --------------ATLTVGGAGAHASYYHKANDQLQVGVEFEASTRMQETSVSFGYQLD 289
Query 378 FRQARV--QGLIDS 389
+A + +G +DS
Sbjct 290 VPKANLLFKGSLDS 303
> hsa:10452 TOMM40, C19orf1, D19S1177E, PER-EC1, PEREC1, TOM40;
translocase of outer mitochondrial membrane 40 homolog (yeast);
K11518 mitochondrial import receptor subunit TOM40
Length=361
Score = 42.4 bits (98), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 57/253 (22%), Positives = 109/253 (43%), Gaps = 40/253 (15%)
Query 147 FDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSYQFGPTVAISPPPDPSQGENAQPPPP 206
+G +L K ++ + Q H+ L T + ES +Y FG T + P++ A P
Sbjct 98 MEGVKLTVNKGLSNHFQVNHTVALST-IGES--NYHFGVTYVGTKQLSPTE---AFP--- 148
Query 207 HFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNT--GRDWASS 264
+ + GSL ++I L K+ +++ +Q F +D G D+ ++
Sbjct 149 --VLVGDMDNSGSLNAQVIHQLGPGLRSKM----AIQTQQSKFVNWQVDGEYRGSDFTAA 202
Query 265 LKV----AWQSCWILDGMFSQVLTPRLQAGCEITYVGANG--ASMLAVGGRYNLDKQSVV 318
+ + IL + Q +TP L G E+ Y G +++++ G+Y L+ +
Sbjct 203 VTLGNPDVLVGSGILVAHYLQSITPCLALGGELVYHRRPGEEGTVMSLAGKYTLNNW-LA 261
Query 319 SCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPETESALRVGWEYLF 378
+ L Q AG+ Y K +D+L + EFE S ++++ G++
Sbjct 262 TVTLGQ-------AGM-------HATYYHKASDQLQVGVEFEASTRMQDTSVSFGYQLDL 307
Query 379 RQARV--QGLIDS 389
+A + +G +DS
Sbjct 308 PKANLLFKGSVDS 320
> mmu:53333 Tomm40, AW539759, MGC118268, Mom35, Tom40; translocase
of outer mitochondrial membrane 40 homolog (yeast); K11518
mitochondrial import receptor subunit TOM40
Length=361
Score = 41.6 bits (96), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 54/237 (22%), Positives = 100/237 (42%), Gaps = 38/237 (16%)
Query 147 FDGFRLEGAKQVNKYLQTCHSFCLGTQLRESGYSYQFGPTVAISPPPDPSQGENAQPPPP 206
+G +L K ++ Q H+ LGT + ES +Y FG T + P++ A P
Sbjct 98 MEGVKLTVNKGLSNRFQVTHTVALGT-IGES--NYHFGVTYVGTKQLSPTE---AFP--- 148
Query 207 HFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNT--GRDWASS 264
+ + GSL ++I L K+ +++ +Q F +D G D+ ++
Sbjct 149 --VLVGDMDNSGSLNAQVIHQLSPGLRSKM----AIQTQQSKFVNWQVDGEYRGSDFTAA 202
Query 265 LKV----AWQSCWILDGMFSQVLTPRLQAGCEITYVGANG--ASMLAVGGRYNLDKQSVV 318
+ + IL + Q +TP L G E+ Y G +++++ G+Y L+ +
Sbjct 203 VTLGNPDVLVGSGILVAHYLQSITPCLALGGELVYHRRPGEEGTVMSLAGKYTLNNW-LA 261
Query 319 SCQLSQQPDFKSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPETESALRVGWE 375
+ L Q AG+ Y K +D+L + EFE S +++ G++
Sbjct 262 TVTLGQ-------AGM-------HATYYHKASDQLQVGVEFEASTRMQDTSASFGYQ 304
> sce:YMR203W TOM40, ISP42, MOM38; Component of the TOM (translocase
of outer membrane) complex responsible for recognition
and initial import steps for all mitochondrially directed
proteins; constitutes the core element of the protein conducting
pore; K11518 mitochondrial import receptor subunit TOM40
Length=387
Score = 40.4 bits (93), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 70/298 (23%), Positives = 113/298 (37%), Gaps = 49/298 (16%)
Query 147 FDGFR--LEGAKQVNKYLQTCHSFCLGTQ-LRESGYSYQFGPTVAISPPPDPSQGENAQP 203
F G R L A +N QT H+F +G+Q L + +S F
Sbjct 81 FTGLRADLNKAFSMNPAFQTSHTFSIGSQALPKYAFSALFAND----------------- 123
Query 204 PPPHFFAMAKLGTDGSLQGRLIKTLCQSADLKLNFNSSLKDEQRNFYEISLDNTGRDWAS 263
+ FA + D S+ GRL + K+N + D Q ++ D D++
Sbjct 124 ---NLFAQGNIDNDLSVSGRLNYGWDKKNISKVNLQ--ISDGQPTMCQLEQDYQASDFSV 178
Query 264 SLKVAWQS-------CWILDGMFSQVLTPRLQAGCEITYVGANGASMLAVG----GRYNL 312
++K S + F Q +TP+L G E Y +G++ G RY
Sbjct 179 NVKTLNPSFSEKGEFTGVAVASFLQSVTPQLALGLETLYSRTDGSAPGDAGVSYLTRYVS 238
Query 313 DKQS-VVSCQLSQQPDF------KSPAGLTKLTHTCKVQYARKVTDRLSMASEFEYSHPE 365
KQ + S QL K + T +TD L M + P
Sbjct 239 KKQDWIFSGQLQANGALIASLWRKVAQNVEAGIETTLQAGMVPITDPL-MGTPIGI-QPT 296
Query 366 TESALRVGWEYLFRQARVQGLIDSCGRISVFAQDYNGFGVS----GLIDYWRGTYKFG 419
E + +G +Y +RQ+ +G +DS G+++ F + +S G ID+++ K G
Sbjct 297 VEGSTTIGAKYEYRQSVYRGTLDSNGKVACFLERKVLPTLSVLFCGEIDHFKNDTKIG 354
> tgo:TGME49_023780 hypothetical protein
Length=1883
Score = 34.3 bits (77), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 16/29 (55%), Positives = 18/29 (62%), Gaps = 1/29 (3%)
Query 180 SYQFGPTVAISPPPDPSQGENAQPPP-PH 207
S+ F PTV ISPP P N+ PPP PH
Sbjct 189 SFAFSPTVGISPPSAPVTTHNSSPPPSPH 217
> sce:YIL151C Putative protein of unknown function, predicted
to contain a PINc domain
Length=1118
Score = 32.0 bits (71), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 43/192 (22%), Positives = 78/192 (40%), Gaps = 14/192 (7%)
Query 21 DFLRRGDQRADGCRQRRGESVRGGRSRCASSVSVGKDDSPAVARCSEKSSGFLGVPGLGF 80
D+ R + R S + SS S +D+P + + G + G GF
Sbjct 17 DYSRSNNNHTHIVSDMRPTSAAFLHQKRHSSSS--HNDTPESSFAKRRVPGIVDPVGKGF 74
Query 81 FGGSKESQSASSDAPAAVDKREKARLDTLARE-NEQTPEPPKKPQPTQPIPFE--HFSRE 137
G SQ ++ + P+ D + +R +++R+ E TP+ PT +P + +R
Sbjct 75 IDGITNSQISAQNTPSKTD--DASRRPSISRKVMESTPQVKTSSIPTMDVPKSPYYVNRT 132
Query 138 W----MNISGQDTFDGFRLEGAKQVNKYLQTCHSFCLGTQLRE--SGYSYQFGPTVAISP 191
M + +DT++ + ++ L + +Q + + + P VA SP
Sbjct 133 MLARNMKVVSRDTYED-NANPQMRADEPLVASNGIYSNSQPQSQVTLSDIRRAPVVAASP 191
Query 192 PPDPSQGENAQP 203
PP Q +AQP
Sbjct 192 PPMIRQLPSAQP 203
Lambda K H
0.317 0.134 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 20219742492
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40