bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3104_orf1
Length=195
Score E
Sequences producing significant alignments: (Bits) Value
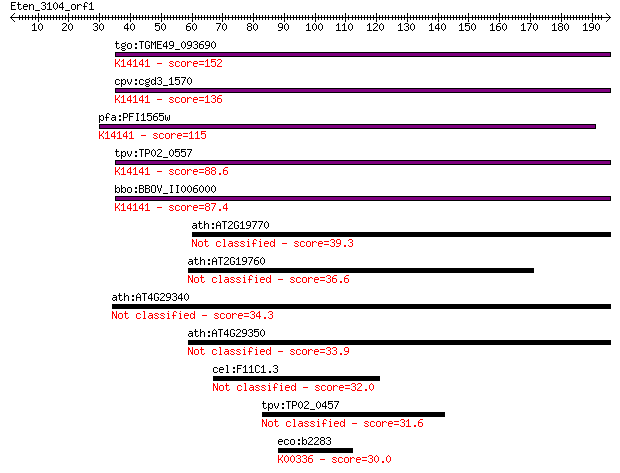
tgo:TGME49_093690 profilin family protein ; K14141 profilin-li... 152 6e-37
cpv:cgd3_1570 sporozoite antigen ; K14141 profilin-like protein 136 5e-32
pfa:PFI1565w PfPfn; profilin, putative; K14141 profilin-like p... 115 1e-25
tpv:TP02_0557 hypothetical protein; K14141 profilin-like protein 88.6 1e-17
bbo:BBOV_II006000 18.m06498; hypothetical protein; K14141 prof... 87.4 3e-17
ath:AT2G19770 PRF5; PRF5 (PROFILIN5); actin binding / actin mo... 39.3 0.009
ath:AT2G19760 PRF1; PRF1 (PROFILIN 1); actin binding 36.6 0.057
ath:AT4G29340 PRF4; PRF4 (PROFILIN 4); actin binding 34.3 0.30
ath:AT4G29350 PFN2; PFN2 (PROFILIN 2); actin binding / protein... 33.9 0.39
cel:F11C1.3 hypothetical protein 32.0 1.5
tpv:TP02_0457 hypothetical protein 31.6 1.9
eco:b2283 nuoG, ECK2277, JW2278; NADH:ubiquinone oxidoreductas... 30.0 5.4
> tgo:TGME49_093690 profilin family protein ; K14141 profilin-like
protein
Length=163
Score = 152 bits (384), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 79/162 (48%), Positives = 111/162 (68%), Gaps = 3/162 (1%)
Query 35 WDTSVREWLVDTGRVFAGGVASIADGCRLFGAAVEGEGNAWEELVKTNYQIEVPQEDGTS 94
WD V+EWLVDTG AGG+A+ DG AA + +G W +L K +++ + EDG +
Sbjct 4 WDPVVKEWLVDTGYCCAGGIANAEDGVVFAAAADDDDG--WSKLYKDDHEEDTIGEDGNA 61
Query 95 I-SVDCDEAETLRQAVVDGRAPNGVYIGGTKYKLAEVKRDFTFNDQNYDVAILGKNKGGG 153
V +EA T++ AV DG APNGV+IGG KYK+ ++ F +ND +D+ + ++KGG
Sbjct 62 CGKVSINEASTIKAAVDDGSAPNGVWIGGQKYKVVRPEKGFEYNDCTFDITMCARSKGGA 121
Query 154 FLIKTPNENVVIALYDEEKEQNKADALTTALNFAEYLYQGGF 195
LIKTPN ++VIALYDEEKEQ+K ++ T+AL FAEYL+Q G+
Sbjct 122 HLIKTPNGSIVIALYDEEKEQDKGNSRTSALAFAEYLHQSGY 163
> cpv:cgd3_1570 sporozoite antigen ; K14141 profilin-like protein
Length=162
Score = 136 bits (342), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 72/162 (44%), Positives = 103/162 (63%), Gaps = 4/162 (2%)
Query 35 WDTSVREWLVDTGRVFAGGVASIADGCRLFGAAVEGEGNAWEELVKTNYQIEVPQEDGTS 94
WD V+EWL+DTG V AGG+ SI DG F AA +G+AW+ LV+ +++ V Q DG S
Sbjct 4 WDDMVKEWLIDTGSVCAGGLCSI-DG--AFYAASADQGDAWKTLVREDHEENVIQSDGVS 60
Query 95 ISVD-CDEAETLRQAVVDGRAPNGVYIGGTKYKLAEVKRDFTFNDQNYDVAILGKNKGGG 153
+ + ++ TL QA+ +G+APNGV++GG KYK+ V++DF ND V + +GG
Sbjct 61 EAAELINDQTTLCQAISEGKAPNGVWVGGNKYKIIRVEKDFQQNDATVHVTFCNRPQGGC 120
Query 154 FLIKTPNENVVIALYDEEKEQNKADALTTALNFAEYLYQGGF 195
FL+ T N VV+A+YDE K+Q+ + AL AEYL G+
Sbjct 121 FLVDTQNGTVVVAVYDESKDQSSGNCKKVALQLAEYLVSQGY 162
> pfa:PFI1565w PfPfn; profilin, putative; K14141 profilin-like
protein
Length=171
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 62/165 (37%), Positives = 95/165 (57%), Gaps = 4/165 (2%)
Query 30 ADTQAWDTSVREWLVDTGRVFAGGVASIADG----CRLFGAAVEGEGNAWEELVKTNYQI 85
A+ +WD+ + + L+ T +V G+AS DG C G + + W K +Y I
Sbjct 2 AEEYSWDSYLNDRLLATNQVSGAGLASEEDGVVYACVAQGEESDPNFDKWSLFYKEDYDI 61
Query 86 EVPQEDGTSISVDCDEAETLRQAVVDGRAPNGVYIGGTKYKLAEVKRDFTFNDQNYDVAI 145
EV E+GT + +E +T+ +G AP+GV++GGTKY+ ++RD F N+DVA
Sbjct 62 EVEDENGTKTTKTINEGQTILVVFNEGYAPDGVWLGGTKYQFINIERDLEFEGYNFDVAT 121
Query 146 LGKNKGGGFLIKTPNENVVIALYDEEKEQNKADALTTALNFAEYL 190
K KGG L+K P N+++ LYDEEKEQ++ ++ AL FA+ L
Sbjct 122 CAKLKGGLHLVKVPGGNILVVLYDEEKEQDRGNSKIAALTFAKEL 166
> tpv:TP02_0557 hypothetical protein; K14141 profilin-like protein
Length=164
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 77/161 (47%), Gaps = 0/161 (0%)
Query 35 WDTSVREWLVDTGRVFAGGVASIADGCRLFGAAVEGEGNAWEELVKTNYQIEVPQEDGTS 94
W ++E + + G+A+ DG + ++ EG W+ + K Y E + G
Sbjct 4 WVPVLKETALSNNSCYGAGIANGEDGELFVASDLDHEGLHWDSVYKDPYVYETFDDAGNP 63
Query 95 ISVDCDEAETLRQAVVDGRAPNGVYIGGTKYKLAEVKRDFTFNDQNYDVAILGKNKGGGF 154
+ ++ DE T+R+ + G+++GG KY A D ++ KNKGG
Sbjct 64 LKINVDEKFTIREVFEKKMSSEGIFLGGEKYTFASYDPDMESGSFKFECVCGAKNKGGCH 123
Query 155 LIKTPNENVVIALYDEEKEQNKADALTTALNFAEYLYQGGF 195
LIKTP +V+ +YDE + Q+K + A N AEYL G+
Sbjct 124 LIKTPGNYIVVVVYDETRGQDKTVSRMAAFNLAEYLATNGY 164
> bbo:BBOV_II006000 18.m06498; hypothetical protein; K14141 profilin-like
protein
Length=164
Score = 87.4 bits (215), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 48/162 (29%), Positives = 78/162 (48%), Gaps = 2/162 (1%)
Query 35 WDTSVREWLVDTGRVFAGGVASIADGCRLFGAA-VEGEGNAWEELVKTNYQIEVPQEDGT 93
W ++++ + + G+A+ DG LF AA ++ + W+ + + Y+ E E+G
Sbjct 4 WVPTIKQLALADNACYGCGIANAEDG-ELFSAADIDHDDLCWDSVYRDPYEFEATDENGQ 62
Query 94 SISVDCDEAETLRQAVVDGRAPNGVYIGGTKYKLAEVKRDFTFNDQNYDVAILGKNKGGG 153
I E T+ + R+ G++IGG KY A D D + KNKGG
Sbjct 63 PIKHQITEKATIMEVFEKRRSSIGIFIGGNKYTFANYDDDCPVGDYTFKCVSAAKNKGGA 122
Query 154 FLIKTPNENVVIALYDEEKEQNKADALTTALNFAEYLYQGGF 195
L+KTP +VI ++DE + QNK + A AEY+ G+
Sbjct 123 HLVKTPGGYIVICVFDENRGQNKTASRMAAFALAEYMAANGY 164
> ath:AT2G19770 PRF5; PRF5 (PROFILIN5); actin binding / actin
monomer binding
Length=134
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 38/137 (27%), Positives = 59/137 (43%), Gaps = 23/137 (16%)
Query 60 GCRLFGAAVEG-EGNAWEELVKTNYQIEVPQEDGTSISVDCDEAETLRQAVVDGRAPNGV 118
G L AA+ G +G+ W + N+ PQE T I D DE L AP G+
Sbjct 20 GHHLTAAAIIGHDGSVWAQ--SANFPQFKPQEI-TDIMKDFDEPGHL--------APTGM 68
Query 119 YIGGTKYKLAEVKRDFTFNDQNYDVAILGKNKGGGFLIKTPNENVVIALYDEEKEQNKAD 178
++ G KY + + + + I GK GG IK +++V LY+E + +
Sbjct 69 FLAGLKYMVIQGEPN---------AVIRGKKGAGGITIKKTGQSMVFGLYEEPVTPGQCN 119
Query 179 ALTTALNFAEYLYQGGF 195
+ L +YL + G
Sbjct 120 MVVERL--GDYLIEQGL 134
> ath:AT2G19760 PRF1; PRF1 (PROFILIN 1); actin binding
Length=131
Score = 36.6 bits (83), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 52/114 (45%), Gaps = 23/114 (20%)
Query 59 DGCRLFGAAVEGE-GNAWEELVKTNYQIEVPQE-DGTSISVDCDEAETLRQAVVDGRAPN 116
+G L AA+ G+ G+ W + K + PQE DG I D +E L AP
Sbjct 16 EGNHLTAAAILGQDGSVWAQSAK--FPQLKPQEIDG--IKKDFEEPGFL--------APT 63
Query 117 GVYIGGTKYKLAEVKRDFTFNDQNYDVAILGKNKGGGFLIKTPNENVVIALYDE 170
G+++GG KY + + ++ I GK GG IK N+ +V YDE
Sbjct 64 GLFLGGEKYMVIQGEQG---------AVIRGKKGPGGVTIKKTNQALVFGFYDE 108
> ath:AT4G29340 PRF4; PRF4 (PROFILIN 4); actin binding
Length=134
Score = 34.3 bits (77), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 43/167 (25%), Positives = 66/167 (39%), Gaps = 39/167 (23%)
Query 34 AWDTSVREWLV-DTGRVFAGGVASIADGCRLFGAAVEG-EGNAWEELVKTNYQIEVPQED 91
+W T V E L+ D G G L AA+ G +G+ W + N+ PQ
Sbjct 2 SWQTYVDEHLMCDVGD---------GQGHHLTAAAIVGHDGSVWAQ--SANF----PQFK 46
Query 92 G---TSISVDCDEAETLRQAVVDGRAPNGVYIGGTKYKLAEVKRDFTFNDQNYDVAILGK 148
G + I D DE L AP G+++ G KY + + + I GK
Sbjct 47 GQEFSDIMKDFDEPGHL--------APTGLFMAGAKYMVIQGEPG---------AVIRGK 89
Query 149 NKGGGFLIKTPNENVVIALYDEEKEQNKADALTTALNFAEYLYQGGF 195
GG IK ++ V +Y+E + + + L +YL + G
Sbjct 90 KGAGGITIKKTGQSCVFGIYEEPVTPGQCNMVVERL--GDYLLEQGL 134
> ath:AT4G29350 PFN2; PFN2 (PROFILIN 2); actin binding / protein
binding
Length=131
Score = 33.9 bits (76), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 36/138 (26%), Positives = 59/138 (42%), Gaps = 23/138 (16%)
Query 59 DGCRLFGAAVEGE-GNAWEELVKTNYQIEVPQEDGTSISVDCDEAETLRQAVVDGRAPNG 117
+G L AA+ G+ G+ W + + + P E I+ D +EA L AP G
Sbjct 16 EGNHLTHAAIFGQDGSVWAQ--SSAFPQLKPAEI-AGINKDFEEAGHL--------APTG 64
Query 118 VYIGGTKYKLAEVKRDFTFNDQNYDVAILGKNKGGGFLIKTPNENVVIALYDEEKEQNKA 177
+++GG KY + + + I GK GG IK + +V +YDE +
Sbjct 65 LFLGGEKYMVVQGEAG---------AVIRGKKGPGGVTIKKTTQALVFGIYDEPMTGGQC 115
Query 178 DALTTALNFAEYLYQGGF 195
+ + L +YL + G
Sbjct 116 NLVVERL--GDYLIESGL 131
> cel:F11C1.3 hypothetical protein
Length=560
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 7/61 (11%)
Query 67 AVEGEGNAWEELVKTNYQIEVP-------QEDGTSISVDCDEAETLRQAVVDGRAPNGVY 119
A++ G +E K NY P Q +G VDC++ + A DG NG Y
Sbjct 329 AIKNPGYRYENFEKVNYFPNWPCGLNHTKQNNGNCALVDCNQYDNFCNACCDGSHVNGTY 388
Query 120 I 120
+
Sbjct 389 V 389
> tpv:TP02_0457 hypothetical protein
Length=816
Score = 31.6 bits (70), Expect = 1.9, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 35/67 (52%), Gaps = 8/67 (11%)
Query 83 YQI-EVPQEDGT---SISVD-CDEAETLRQAVVDGR---APNGVYIGGTKYKLAEVKRDF 134
Y+I + P+ D T +IS+D C + + + +VD +PN + I Y++ + R+
Sbjct 713 YEITDTPKSDTTQSDNISMDFCKDCNKVVECIVDVEYLSSPNILIIDFINYRVENIPREM 772
Query 135 TFNDQNY 141
FND Y
Sbjct 773 DFNDHKY 779
> eco:b2283 nuoG, ECK2277, JW2278; NADH:ubiquinone oxidoreductase,
chain G (EC:1.6.5.3); K00336 NADH dehydrogenase I subunit
G [EC:1.6.5.3]
Length=908
Score = 30.0 bits (66), Expect = 5.4, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 88 PQEDGTSISVDCDEAETLRQAVVD 111
P DGT IS+D +EA+ R++VV+
Sbjct 70 PASDGTFISIDDEEAKQFRESVVE 93
Lambda K H
0.317 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5673387808
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40