bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3074_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
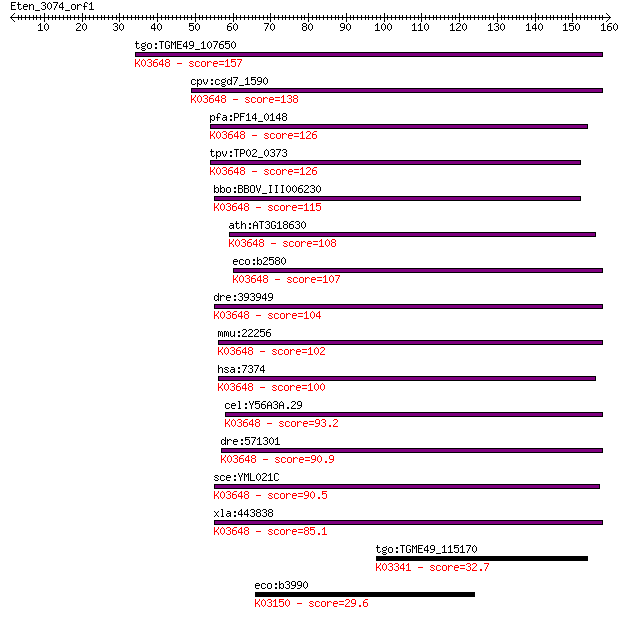
tgo:TGME49_107650 uracil-DNA glycosylase, putative ; K03648 ur... 157 2e-38
cpv:cgd7_1590 uracil-DNA glycosylase ; K03648 uracil-DNA glyco... 138 7e-33
pfa:PF14_0148 uracil-DNA glycosylase, putative (EC:3.2.2.-); K... 126 2e-29
tpv:TP02_0373 uracil-DNA glycosylase; K03648 uracil-DNA glycos... 126 3e-29
bbo:BBOV_III006230 17.m07553; uracil-DNA glycosylase family pr... 115 4e-26
ath:AT3G18630 uracil DNA glycosylase family protein; K03648 ur... 108 8e-24
eco:b2580 ung, ECK2578, JW2564; uracil-DNA-glycosylase (EC:3.2... 107 1e-23
dre:393949 ung, MGC56102, zgc:56102; uracil-DNA glycosylase (E... 104 1e-22
mmu:22256 Ung, UNG1, UNG2; uracil DNA glycosylase (EC:3.2.2.27... 102 7e-22
hsa:7374 UNG, DGU, DKFZp781L1143, HIGM4, UDG, UNG1, UNG15, UNG... 100 3e-21
cel:Y56A3A.29 ung-1; Uracil DNA N-Glycosylase family member (u... 93.2 4e-19
dre:571301 si:dkey-202g15.9; K03648 uracil-DNA glycosylase [EC... 90.9 2e-18
sce:YML021C UNG1; Ung1p (EC:3.2.2.-); K03648 uracil-DNA glycos... 90.5 2e-18
xla:443838 ung, MGC82673; uracil-DNA glycosylase; K03648 uraci... 85.1 9e-17
tgo:TGME49_115170 soluble liver antigen/liver pancreas antigen... 32.7 0.50
eco:b3990 thiH, ECK3981, JW3953, thiB; tyrosine lyase, involve... 29.6 4.2
> tgo:TGME49_107650 uracil-DNA glycosylase, putative ; K03648
uracil-DNA glycosylase [EC:3.2.2.-]
Length=340
Score = 157 bits (396), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 70/124 (56%), Positives = 93/124 (75%), Gaps = 2/124 (1%)
Query 34 ASAVSPFGTFVSTAAAEKMWREAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIY 93
A V P G +T+ A +WR+ +GD W++AL+ EL+ PYFR C+ ++++R+K+ +Y
Sbjct 87 ARLVDPEGEMKATSEAAALWRQTLGDCWFDALKEELRLPYFRDCMQFIRQERQKY--RVY 144
Query 94 PPQELMFKAFRSTPLDKVKVVIVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDR 153
PP L+F AF+ TPL+ VKVV+VGQDPYHQPGQAMGL FSVPRG+ PPSL NIFKEI R
Sbjct 145 PPSRLVFNAFKQTPLNAVKVVVVGQDPYHQPGQAMGLAFSVPRGIPLPPSLQNIFKEIAR 204
Query 154 SIPG 157
+ PG
Sbjct 205 NYPG 208
> cpv:cgd7_1590 uracil-DNA glycosylase ; K03648 uracil-DNA glycosylase
[EC:3.2.2.-]
Length=211
Score = 138 bits (348), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 64/109 (58%), Positives = 82/109 (75%), Gaps = 2/109 (1%)
Query 49 AEKMWREAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPL 108
+E RE GD W+EAL+ EL+KPYF KC+ V+E+R + +YPP + MF F++TPL
Sbjct 63 SESELREYFGDEWFEALKDELRKPYFVKCMKKVQERRNY--AKVYPPSDKMFSCFKATPL 120
Query 109 DKVKVVIVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
+K+ VVI+GQDPYHQPGQAMGL FSVP+GV PPSL NI+KEI + PG
Sbjct 121 NKISVVILGQDPYHQPGQAMGLSFSVPKGVPVPPSLRNIYKEIGKGNPG 169
> pfa:PF14_0148 uracil-DNA glycosylase, putative (EC:3.2.2.-);
K03648 uracil-DNA glycosylase [EC:3.2.2.-]
Length=322
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 57/100 (57%), Positives = 72/100 (72%), Gaps = 2/100 (2%)
Query 54 REAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKV 113
++ M WYE L+ EL+K YF+ L +KE+R+ IYPP++L+F AF TPL +KV
Sbjct 106 KKLMHIEWYELLKDELKKNYFKNMYLKIKEERK--TKVIYPPEQLVFNAFLKTPLSNIKV 163
Query 114 VIVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDR 153
VIVGQDPYHQ QAMGLCFSVP GV+ PPSL NI KE+ +
Sbjct 164 VIVGQDPYHQKDQAMGLCFSVPIGVKIPPSLKNILKEMKQ 203
> tpv:TP02_0373 uracil-DNA glycosylase; K03648 uracil-DNA glycosylase
[EC:3.2.2.-]
Length=286
Score = 126 bits (317), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 59/98 (60%), Positives = 68/98 (69%), Gaps = 2/98 (2%)
Query 54 REAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKV 113
R +GD W+E L E+ KPYF+ L K E+ IYPP L+F AF+ TPL K+KV
Sbjct 70 RNMLGDEWFEVLDSEINKPYFKS--LWNKVLNERSSKKIYPPAHLVFNAFKLTPLSKIKV 127
Query 114 VIVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEI 151
VIVGQDPYHQP QAMGLCFSVP+GV PPSL NI EI
Sbjct 128 VIVGQDPYHQPRQAMGLCFSVPKGVLLPPSLKNILSEI 165
> bbo:BBOV_III006230 17.m07553; uracil-DNA glycosylase family
protein; K03648 uracil-DNA glycosylase [EC:3.2.2.-]
Length=287
Score = 115 bits (289), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 55/97 (56%), Positives = 68/97 (70%), Gaps = 2/97 (2%)
Query 55 EAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVV 114
+ +G+ W + L E++KPYF V + R+ +YPP+ L+F AF+ PL KVKVV
Sbjct 72 KLLGEEWSDKLNNEIKKPYFGNLWAKVNKDRKN--KRVYPPEHLVFNAFQIVPLSKVKVV 129
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEI 151
IVGQDPYHQP QAMGL FSVPRGV PPSL NI+KEI
Sbjct 130 IVGQDPYHQPKQAMGLSFSVPRGVAIPPSLRNIYKEI 166
> ath:AT3G18630 uracil DNA glycosylase family protein; K03648
uracil-DNA glycosylase [EC:3.2.2.-]
Length=330
Score = 108 bits (270), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 54/101 (53%), Positives = 67/101 (66%), Gaps = 6/101 (5%)
Query 59 DSWYEALQGELQKPYFRKCLLAVKEQREKFPSS----IYPPQELMFKAFRSTPLDKVKVV 114
+SW +AL GE KPY + L+ +RE S IYPPQ L+F A +TP D+VK V
Sbjct 111 ESWLKALPGEFHKPYAKS--LSDFLEREIITDSKSPLIYPPQHLIFNALNTTPFDRVKTV 168
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSI 155
I+GQDPYH PGQAMGL FSVP G + P SL NIFKE+ + +
Sbjct 169 IIGQDPYHGPGQAMGLSFSVPEGEKLPSSLLNIFKELHKDV 209
> eco:b2580 ung, ECK2578, JW2564; uracil-DNA-glycosylase (EC:3.2.2.-);
K03648 uracil-DNA glycosylase [EC:3.2.2.-]
Length=229
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 52/98 (53%), Positives = 68/98 (69%), Gaps = 1/98 (1%)
Query 60 SWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVVIVGQD 119
+W++ L E Q+PYF L V +R+ +IYPPQ+ +F AFR T L VKVVI+GQD
Sbjct 6 TWHDVLAEEKQQPYFLNTLQTVASERQS-GVTIYPPQKDVFNAFRFTELGDVKVVILGQD 64
Query 120 PYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
PYH PGQA GL FSV G+ PPSL N++KE++ +IPG
Sbjct 65 PYHGPGQAHGLAFSVRPGIAIPPSLLNMYKELENTIPG 102
> dre:393949 ung, MGC56102, zgc:56102; uracil-DNA glycosylase
(EC:3.2.2.3); K03648 uracil-DNA glycosylase [EC:3.2.2.-]
Length=291
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 52/103 (50%), Positives = 67/103 (65%), Gaps = 2/103 (1%)
Query 55 EAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVV 114
+ +G+SW +AL E K YF+ + V E+R+K +IYPP +F ++ + VKVV
Sbjct 70 DGIGESWLKALSAEFGKSYFKSLMSFVGEERKKH--TIYPPPHAVFTWTQTCDIKDVKVV 127
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
I+GQDPYH P QA GLCFSV R V PPPSL NIFKE+ I G
Sbjct 128 ILGQDPYHGPNQAHGLCFSVQRPVPPPPSLVNIFKELASDIEG 170
> mmu:22256 Ung, UNG1, UNG2; uracil DNA glycosylase (EC:3.2.2.27);
K03648 uracil-DNA glycosylase [EC:3.2.2.-]
Length=306
Score = 102 bits (253), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 63/102 (61%), Gaps = 2/102 (1%)
Query 56 AMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVVI 115
G+SW + L GE KPYF K + V E+R +YPP E +F + + VKVVI
Sbjct 86 GFGESWKQQLCGEFGKPYFVKLMGFVAEERNHH--KVYPPPEQVFTWTQMCDIRDVKVVI 143
Query 116 VGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
+GQDPYH P QA GLCFSV R V PPPSL NIFKE+ I G
Sbjct 144 LGQDPYHGPNQAHGLCFSVQRPVPPPPSLENIFKELSTDIDG 185
> hsa:7374 UNG, DGU, DKFZp781L1143, HIGM4, UDG, UNG1, UNG15, UNG2;
uracil-DNA glycosylase (EC:3.2.2.27); K03648 uracil-DNA
glycosylase [EC:3.2.2.-]
Length=304
Score = 100 bits (248), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 50/100 (50%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 56 AMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVVI 115
G+SW + L GE KPYF K + V E+R+ + ++YPP +F + + VKVVI
Sbjct 84 GFGESWKKHLSGEFGKPYFIKLMGFVAEERKHY--TVYPPPHQVFTWTQMCDIKDVKVVI 141
Query 116 VGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSI 155
+GQDPYH P QA GLCFSV R V PPPSL NI+KE+ I
Sbjct 142 LGQDPYHGPNQAHGLCFSVQRPVPPPPSLENIYKELSTDI 181
> cel:Y56A3A.29 ung-1; Uracil DNA N-Glycosylase family member
(ung-1); K03648 uracil-DNA glycosylase [EC:3.2.2.-]
Length=282
Score = 93.2 bits (230), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 47/107 (43%), Positives = 63/107 (58%), Gaps = 15/107 (14%)
Query 58 GDSWYEALQGELQKPYFRKCLLAVKEQREKFPSS-------IYPPQELMFKAFRSTPLDK 110
G+SW + L+ E +K Y K EKF +S ++PP +F F P D+
Sbjct 63 GESWSKLLEEEFKKGYISKI--------EKFLNSEVNKGKQVFPPPTQIFTTFNLLPFDE 114
Query 111 VKVVIVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
+ VVI+GQDPYH QA GL FSV +GV+PPPSL NI+KE++ I G
Sbjct 115 ISVVIIGQDPYHDDNQAHGLSFSVQKGVKPPPSLKNIYKELESDIEG 161
> dre:571301 si:dkey-202g15.9; K03648 uracil-DNA glycosylase [EC:3.2.2.-]
Length=250
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/101 (47%), Positives = 61/101 (60%), Gaps = 2/101 (1%)
Query 57 MGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVVIV 116
+G+SW + E KPYF K + V +R+ S++YP E +F ++ VKVVI+
Sbjct 31 VGESWRRHIGTEFAKPYFTKLMSFVTIERK--CSTVYPSLEQVFYWTTLCAIEDVKVVIL 88
Query 117 GQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
GQDPYH PGQA GL FSV R PPPSL NIF E+ I G
Sbjct 89 GQDPYHHPGQAHGLAFSVLRPKSPPPSLENIFMELQEDIDG 129
> sce:YML021C UNG1; Ung1p (EC:3.2.2.-); K03648 uracil-DNA glycosylase
[EC:3.2.2.-]
Length=359
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 46/102 (45%), Positives = 62/102 (60%), Gaps = 2/102 (1%)
Query 55 EAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVV 114
E + DSW+ L E +KPYF K V +E+ +++PP + ++ R TP +KVKVV
Sbjct 100 ETIDDSWFPHLMDEFKKPYFVKLKQFV--TKEQADHTVFPPAKDIYSWTRLTPFNKVKVV 157
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIP 156
I+GQDPYH QA GL FSV PPSL NI+KE+ + P
Sbjct 158 IIGQDPYHNFNQAHGLAFSVKPPTPAPPSLKNIYKELKQEYP 199
> xla:443838 ung, MGC82673; uracil-DNA glycosylase; K03648 uracil-DNA
glycosylase [EC:3.2.2.-]
Length=304
Score = 85.1 bits (209), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 49/103 (47%), Positives = 66/103 (64%), Gaps = 2/103 (1%)
Query 55 EAMGDSWYEALQGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVV 114
+G+SW + L E KPYF K + E+R+K ++YPP E +F + + VKVV
Sbjct 83 HGLGESWKQELLAEFAKPYFVKLSNFIAEERKK--CTVYPPPEEVFTWTQMVDIKDVKVV 140
Query 115 IVGQDPYHQPGQAMGLCFSVPRGVRPPPSLTNIFKEIDRSIPG 157
I+GQDPYH P QA GLCFSV + V PPPSL N++KE++ I G
Sbjct 141 ILGQDPYHGPNQAHGLCFSVKKPVPPPPSLVNMYKELETDIEG 183
> tgo:TGME49_115170 soluble liver antigen/liver pancreas antigen
domain-containing protein (EC:4.1.1.25); K03341 O-phospho-L-seryl-tRNASec:L-selenocysteinyl-tRNA
synthase [EC:2.9.1.2]
Length=656
Score = 32.7 bits (73), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 28/61 (45%), Gaps = 5/61 (8%)
Query 98 LMFKAFRSTPLDKVKVVIVGQDPYHQP-----GQAMGLCFSVPRGVRPPPSLTNIFKEID 152
L+F R LD + + + P G ++ LCFS R VRPP + IF ID
Sbjct 153 LLFALTRRLVLDAIHICGIQAARAALPVPFATGLSLTLCFSALRTVRPPSARFIIFSRID 212
Query 153 R 153
+
Sbjct 213 Q 213
> eco:b3990 thiH, ECK3981, JW3953, thiB; tyrosine lyase, involved
in thiamin-thiazole moiety synthesis; K03150 thiamine biosynthesis
ThiH
Length=377
Score = 29.6 bits (65), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 28/58 (48%), Gaps = 3/58 (5%)
Query 66 QGELQKPYFRKCLLAVKEQREKFPSSIYPPQELMFKAFRSTPLDKVKVVIVGQDPYHQ 123
Q ++ YFR+ L A++EQ + P E + + LD V+V Q+ YH+
Sbjct 133 QAKVGMDYFRRHLPALREQFSSLQMEVQPLAETEYAELKQLGLDG---VMVYQETYHE 187
Lambda K H
0.319 0.134 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40