bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3068_orf1
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
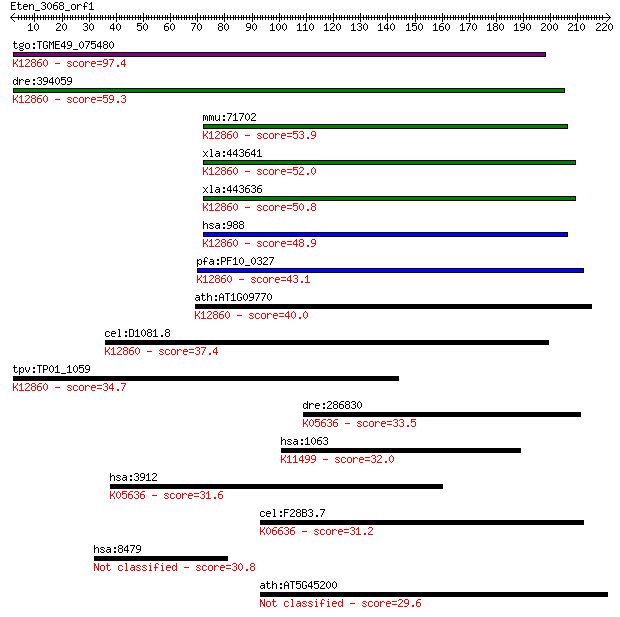
tgo:TGME49_075480 myb-like DNA-binding domain-containing prote... 97.4 4e-20
dre:394059 cdc5l, MGC55853, zgc:55853; CDC5 cell division cycl... 59.3 1e-08
mmu:71702 Cdc5l, 1200002I02Rik, AA408004, PCDC5RP; cell divisi... 53.9 4e-07
xla:443641 cdc5l, MGC114655; cell division cycle 5-like; K1286... 52.0 2e-06
xla:443636 cdc5l, MGC154633; CDC5 cell division cycle 5-like; ... 50.8 3e-06
hsa:988 CDC5L, CDC5, CDC5-LIKE, CEF1, KIAA0432, PCDC5RP, dJ319... 48.9 1e-05
pfa:PF10_0327 Myb2 protein; K12860 pre-mRNA-splicing factor CD... 43.1 7e-04
ath:AT1G09770 ATCDC5; ATCDC5 (ARABIDOPSIS THALIANA CELL DIVISI... 40.0 0.006
cel:D1081.8 hypothetical protein; K12860 pre-mRNA-splicing fac... 37.4 0.037
tpv:TP01_1059 hypothetical protein; K12860 pre-mRNA-splicing f... 34.7 0.30
dre:286830 lamb1a, fc20b05, gup, lamb1, sb:cb95, wu:fc20b05; l... 33.5 0.56
hsa:1063 CENPF, CENF, PRO1779, hcp-1; centromere protein F, 35... 32.0 1.6
hsa:3912 LAMB1, CLM, MGC142015; laminin, beta 1; K05636 lamini... 31.6 2.5
cel:F28B3.7 him-1; High Incidence of Males (increased X chromo... 31.2 3.0
hsa:8479 HIRIP3; HIRA interacting protein 3 30.8
ath:AT5G45200 disease resistance protein (TIR-NBS-LRR class), ... 29.6 8.9
> tgo:TGME49_075480 myb-like DNA-binding domain-containing protein
; K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=888
Score = 97.4 bits (241), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 66/206 (32%), Positives = 107/206 (51%), Gaps = 13/206 (6%)
Query 2 LTENVMSLLVQNDCYLHPFRGISPPPPTGELEPCSLEQMQAVRELL---------ETELA 52
+ E VM LL ND HP +G P +LE + + RE+L ++E+A
Sbjct 643 INEEVMVLLA-NDALKHPIKGGKAPSSVPQLEKLDDDLLHEAREMLRCEAELMQHQSEVA 701
Query 53 S-WNLDENYPRFDKLVLTLETQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLAS 111
+ +LD++ R ++ + +F+P++K +V +L SER + Q E LK + +
Sbjct 702 TEGHLDQD--RVARIFDDAQDAFIFSPSAKKFVLYQSLPGSERAEALKRQFEDLKKQVEA 759
Query 112 AVQRTKKLEKKYKVLTTGYTNRQKELAKAISESSAELLQLDARIASFTKLHEAEQKAVKV 171
R KK EKKY VLT GY + L + + E+ + A +A+F L + E A++
Sbjct 760 TANRVKKAEKKYSVLTKGYETKATSLIERLEETYRDFELQAATLAAFQALRDRELPAIEK 819
Query 172 RTEEKRVEVLRERAKHLFLQQLYGRL 197
R +E++ VLRER KHL++Q+ Y L
Sbjct 820 RKQEQQALVLREREKHLWMQRRYAAL 845
> dre:394059 cdc5l, MGC55853, zgc:55853; CDC5 cell division cycle
5-like (S. pombe); K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=800
Score = 59.3 bits (142), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 54/223 (24%), Positives = 102/223 (45%), Gaps = 20/223 (8%)
Query 2 LTENVMSLLVQNDCYLHPF------RGISPPPPTGE---------LEPCSLEQMQAVREL 46
L + M ++ DC HPF +G++ E E S E+++ +L
Sbjct 574 LIKREMITMIHYDCLHHPFSDAKKTKGVNSSSNNAEHISYLEKTPYEKVSEEELKKAGDL 633
Query 47 L--ETELASWNL---DENYPRFDKLVLTLETQLVFNPASKSYVSSSTLSPSERLQVYAHQ 101
L E E+ + D + ++++ +Q+++ P Y ++ S +R+ +
Sbjct 634 LLQEMEVVKHGMGHGDLSMEAYNQVWEECYSQVLYLPGQSRYTRANLASKKDRIDSLEKK 693
Query 102 VETLKTGLASAVQRTKKLEKKYKVLTTGYTNRQKELAKAISESSAELLQLDARIASFTKL 161
+E + + + +R K+EKK K+L GY +R L K +SE +L Q + + +F +L
Sbjct 694 LEMNRGHMTAEAKRAAKMEKKMKILLGGYQSRAMGLLKQLSEVWDQLEQANLELHTFMEL 753
Query 162 HEAEQKAVKVRTEEKRVEVLRERAKHLFLQQLYGRLTAVKKHL 204
+ E A+ R E R +V R++ + LQQ + L K+ L
Sbjct 754 KKQEDLAIPRRQEALREDVQRQQEREKELQQRFADLMLDKQTL 796
> mmu:71702 Cdc5l, 1200002I02Rik, AA408004, PCDC5RP; cell division
cycle 5-like (S. pombe); K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=802
Score = 53.9 bits (128), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 70/134 (52%), Gaps = 0/134 (0%)
Query 72 TQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYT 131
+Q+++ PA Y ++ S +R++ ++E + + + +R K+EKK K+L GY
Sbjct 666 SQVLYLPAQSRYTRANLASKKDRIESLEKRLEINRGHMTTEAKRAAKMEKKMKILLGGYQ 725
Query 132 NRQKELAKAISESSAELLQLDARIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHLFLQ 191
+R L K +++ ++ Q + +F +L + E A+ R E + +V R++ + LQ
Sbjct 726 SRAMGLMKQLNDLWDQIEQAHLELRTFEELKKHEDSAIPRRLECLKEDVQRQQEREKELQ 785
Query 192 QLYGRLTAVKKHLQ 205
Q Y L K+ LQ
Sbjct 786 QRYADLLMEKETLQ 799
> xla:443641 cdc5l, MGC114655; cell division cycle 5-like; K12860
pre-mRNA-splicing factor CDC5/CEF1
Length=804
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 68/137 (49%), Gaps = 0/137 (0%)
Query 72 TQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYT 131
+Q+++ P Y ++ S +R++ ++E + + +R K+EKK K+L GY
Sbjct 668 SQVLYLPGQGRYTRANLASKKDRIESLEKRLEVNRGHMTGEAKRAAKMEKKLKILLGGYQ 727
Query 132 NRQKELAKAISESSAELLQLDARIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHLFLQ 191
+R L K ++E + Q + + +F +L + E A+ R E + +V R+ + LQ
Sbjct 728 SRAMGLIKQLNEVWDQYEQANLELGTFEELKKHEDIAIPRRIECLKEDVQRQEERERELQ 787
Query 192 QLYGRLTAVKKHLQELL 208
Q + L K+ + +L
Sbjct 788 QRFAELMLEKESYEAIL 804
> xla:443636 cdc5l, MGC154633; CDC5 cell division cycle 5-like;
K12860 pre-mRNA-splicing factor CDC5/CEF1
Length=804
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 68/137 (49%), Gaps = 0/137 (0%)
Query 72 TQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYT 131
+Q+++ P Y ++ S +R++ ++E + + + +R K+EKK K+L GY
Sbjct 668 SQVLYLPGQGRYTRANLASKKDRIESLEKRLEINRGHMTAEAKRAAKMEKKLKILLGGYQ 727
Query 132 NRQKELAKAISESSAELLQLDARIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHLFLQ 191
+R L K ++E + Q + + +F +L E A+ R E + +V R+ + LQ
Sbjct 728 SRAMGLIKQLNEIWDQYEQANLELGTFEELKVHEDTAIPRRIECLKEDVQRQEERERELQ 787
Query 192 QLYGRLTAVKKHLQELL 208
Q + L K+ + +L
Sbjct 788 QRFAELMLEKESYEAIL 804
> hsa:988 CDC5L, CDC5, CDC5-LIKE, CEF1, KIAA0432, PCDC5RP, dJ319D22.1;
CDC5 cell division cycle 5-like (S. pombe); K12860
pre-mRNA-splicing factor CDC5/CEF1
Length=802
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 33/134 (24%), Positives = 68/134 (50%), Gaps = 0/134 (0%)
Query 72 TQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYT 131
+Q+++ P Y ++ S +R++ ++E + + + +R K+EKK K+L GY
Sbjct 666 SQVLYLPGQSRYTRANLASKKDRIESLEKRLEINRGHMTTEAKRAAKMEKKMKILLGGYQ 725
Query 132 NRQKELAKAISESSAELLQLDARIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHLFLQ 191
+R L K +++ ++ Q + +F +L + E A+ R E + +V R++ + LQ
Sbjct 726 SRAMGLMKQLNDLWDQIEQAHLELRTFEELKKHEDSAIPRRLECLKEDVQRQQEREKELQ 785
Query 192 QLYGRLTAVKKHLQ 205
Y L K+ L+
Sbjct 786 HRYADLLLEKETLK 799
> pfa:PF10_0327 Myb2 protein; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=915
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 34/142 (23%), Positives = 63/142 (44%), Gaps = 0/142 (0%)
Query 70 LETQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTG 129
+ ++F P+ +Y ++ +++ + Y ++ E LK + + ++ KKLE KY + T G
Sbjct 770 INKNIIFCPSKNAYRFIEDVNENDKKENYKYKCEKLKNLILNDMEHYKKLENKYDIYTKG 829
Query 130 YTNRQKELAKAISESSAELLQLDARIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHLF 189
Y + K K+ + + LHE E+ R +E++ E +E H
Sbjct 830 YQLKIKSYKKSYDTLFNSYINCINEKEALNVLHENEKIYALTRIKEEKKENKKEIEYHKS 889
Query 190 LQQLYGRLTAVKKHLQELLSDT 211
LQ+ Y L L+E T
Sbjct 890 LQKFYQDLLETNHQLKETCKQT 911
> ath:AT1G09770 ATCDC5; ATCDC5 (ARABIDOPSIS THALIANA CELL DIVISION
CYCLE 5); DNA binding / transcription factor; K12860 pre-mRNA-splicing
factor CDC5/CEF1
Length=844
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 29/146 (19%), Positives = 66/146 (45%), Gaps = 0/146 (0%)
Query 69 TLETQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTT 128
T L++ P +Y SS ++++ + ++E ++ + ++ + ++ KYK T
Sbjct 664 TCVNDLMYFPTRSAYELSSVAGNADKVAAFQEEMENVRKKMEEDEKKAEHMKAKYKTYTK 723
Query 129 GYTNRQKELAKAISESSAELLQLDARIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHL 188
G+ R + + I + + + F L E+ A R + + EV++++
Sbjct 724 GHERRAETVWTQIEATLKQAEIGGTEVECFKALKRQEEMAASFRKKNLQEEVIKQKETES 783
Query 189 FLQQLYGRLTAVKKHLQELLSDTGAQ 214
LQ YG + A+ + +E++ AQ
Sbjct 784 KLQTRYGNMLAMVEKAEEIMVGFRAQ 809
> cel:D1081.8 hypothetical protein; K12860 pre-mRNA-splicing factor
CDC5/CEF1
Length=755
Score = 37.4 bits (85), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 36/165 (21%), Positives = 72/165 (43%), Gaps = 8/165 (4%)
Query 36 SLEQMQAVRELLETELASWNLDENYPRFDKLVLTLETQLVFNP--ASKSYVSSSTLSPSE 93
S E++ A +L++ E E+ P + L+ + Q + + + L E
Sbjct 573 SREELDAAADLIKQEA------ESGPELNSLMWKVVEQCTSEIILSKDKFTRIAILPREE 626
Query 94 RLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYTNRQKELAKAISESSAELLQLDA 153
+++ + + + + +R K+EKK +V GY +L K E + E+ +
Sbjct 627 QMKALNDEFQMYRGWMNQRAKRAAKVEKKLRVKLGGYQAIHDKLCKKYQEVTTEIEMANI 686
Query 154 RIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHLFLQQLYGRLT 198
+F +L E E KA+ R + EV + + LQ++Y +L+
Sbjct 687 EKKTFERLGEHELKAINKRVGRLQQEVTTQETREKDLQKMYSKLS 731
> tpv:TP01_1059 hypothetical protein; K12860 pre-mRNA-splicing
factor CDC5/CEF1
Length=658
Score = 34.7 bits (78), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 40/147 (27%), Positives = 63/147 (42%), Gaps = 13/147 (8%)
Query 2 LTENVMSLLVQNDCYLHPFRGISPPPPTGELEPCSLEQMQAVRELL--ETELASWNLDEN 59
+ E ++LL D HP P E L+ ++ R+L+ E E+ S LD+
Sbjct 520 INEEFVNLLAY-DSVKHPIPHGRPSDQYKPKEYFDLKLIEEARKLIDAEAEILSRGLDDR 578
Query 60 YPRFDKLVLTLE---TQLVFNPASKSYVSSSTLSPSERLQVYAHQVETLKTGLASAVQRT 116
LTLE ++ K Y+ L+ E+ + + L + +R
Sbjct 579 N-------LTLEWKTDNFKYSQLQKRYMPCEKLTLQEQKLAEEMLCKEYEKHLENLSKRI 631
Query 117 KKLEKKYKVLTTGYTNRQKELAKAISE 143
K LE KY V T GY NR++++ K I E
Sbjct 632 KSLENKYNVSTGGYRNREEKILKEIGE 658
> dre:286830 lamb1a, fc20b05, gup, lamb1, sb:cb95, wu:fc20b05;
laminin, beta 1a; K05636 laminin, beta 1
Length=1785
Score = 33.5 bits (75), Expect = 0.56, Method: Composition-based stats.
Identities = 36/116 (31%), Positives = 51/116 (43%), Gaps = 14/116 (12%)
Query 109 LASAVQRTKKLEK-----KYKVLTTGY----TNRQKELAKAISESSAELLQLDARIASFT 159
L++A +R KLE K K L T T ++ E A++E + L + + T
Sbjct 1639 LSNATRRLLKLESDVALLKEKALNTSISANSTEKEAESINALAEQLKKDLDSELKDKYST 1698
Query 160 KLHEAEQKAVKVRTEEKRVEVLRERAKHLFLQ-----QLYGRLTAVKKHLQELLSD 210
QKA V +KR E L+E A++L LQ QL L Q+LL D
Sbjct 1699 VEELITQKAEGVAEAKKRAEKLQEEARNLLLQASEKLQLLKNLEKNYDQNQKLLED 1754
> hsa:1063 CENPF, CENF, PRO1779, hcp-1; centromere protein F,
350/400kDa (mitosin); K11499 centromere protein F
Length=3114
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 45/90 (50%), Gaps = 6/90 (6%)
Query 101 QVETLKTGLASAVQRTKKLEKKYKVLTTGYTNRQKELAKAISESSAELLQLDARIASFTK 160
+VETLKT + + K E L + N L K I E +L +LD ++SF
Sbjct 2183 EVETLKTQIEEMARSLKVFELDLVTLRSEKEN----LTKQIQEKQGQLSELDKLLSSFKS 2238
Query 161 -LHEAEQKAVKVRTEEKR-VEVLRERAKHL 188
L E EQ ++++ E K VE+L+ + K L
Sbjct 2239 LLEEKEQAEIQIKEESKTAVEMLQNQLKEL 2268
> hsa:3912 LAMB1, CLM, MGC142015; laminin, beta 1; K05636 laminin,
beta 1
Length=1786
Score = 31.6 bits (70), Expect = 2.5, Method: Composition-based stats.
Identities = 29/122 (23%), Positives = 56/122 (45%), Gaps = 19/122 (15%)
Query 38 EQMQAVRELLETELASWNLDENYPRFDKLVLTLETQLVFNPASKSYVSSSTLSPSERLQV 97
+ + V++ L+ EL DE Y + + L+ A K+ S+ +E LQ
Sbjct 1681 QSAEDVKKTLDGEL-----DEKYKKVENLI-----------AKKTEESADARRKAEMLQ- 1723
Query 98 YAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYTNRQKELAKAISESSAELLQLDARIAS 157
++ +TL S +Q K LE+KY+ ++ +ELA+ E + L + ++A
Sbjct 1724 --NEAKTLLAQANSKLQLLKDLERKYEDNQRYLEDKAQELARLEGEVRSLLKDISQKVAV 1781
Query 158 FT 159
++
Sbjct 1782 YS 1783
> cel:F28B3.7 him-1; High Incidence of Males (increased X chromosome
loss) family member (him-1); K06636 structural maintenance
of chromosome 1
Length=1262
Score = 31.2 bits (69), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 33/138 (23%), Positives = 61/138 (44%), Gaps = 26/138 (18%)
Query 93 ERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYTNRQKELA---KAISESSAELL 149
ER++ V ++T +A+ QR K+ E++ K+L + ++ A +E + EL
Sbjct 429 ERVKAKEGDVRRIETQIATLAQRIKETEEETKILKADLKKIENDVVIDKSAAAEYNKEL- 487
Query 150 QLDARIASFTKLHEAEQKAVKVRTEEKRVEVLRERAKHLFLQQLYGRLTA---------- 199
+A +L EA + + ++R E L E K F + +YGRL
Sbjct 488 -----VAVVRQLSEASGDSAEGERNQRRTEAL-EGLKKNFPESVYGRLVDLCQPSHKRFN 541
Query 200 ------VKKHLQELLSDT 211
++KH+ ++ DT
Sbjct 542 IATTKILQKHMNSIVCDT 559
> hsa:8479 HIRIP3; HIRA interacting protein 3
Length=556
Score = 30.8 bits (68), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 27/49 (55%), Gaps = 3/49 (6%)
Query 32 LEPCSLEQMQAVRELLETELASWNLDENYPRFDKLVLTLETQLVFNPAS 80
LEP E+ QA++ L+E EL +DE R DKL LT + + P S
Sbjct 42 LEP---EEKQALKRLVEEELLKMQVDEAASREDKLDLTKKGKRPPTPCS 87
> ath:AT5G45200 disease resistance protein (TIR-NBS-LRR class),
putative
Length=1261
Score = 29.6 bits (65), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 59/135 (43%), Gaps = 13/135 (9%)
Query 93 ERLQVYAHQVETLKTGLASAVQRTKKLEKKYKVLTTGYTNRQ---KELAKAISESSAELL 149
ER+ V+ ET+ TGL + QR ++ + V+++ YT Q EL K A
Sbjct 45 ERINVFIDTRETMGTGLENLFQRIQESKIAIVVISSRYTESQWCLNELVKIKECVEA--- 101
Query 150 QLDARIASFTKLHEAEQKAVKVRT----EEKRVEVLRERAKHLFLQQLYGRLTAVKKHLQ 205
+ F ++ + K V+ T E+ VLR ++ +Q +T+
Sbjct 102 ---GTLVVFPVFYKVDVKIVRFLTGSFGEKLETLVLRHSERYEPWKQALEFVTSKTGKRV 158
Query 206 ELLSDTGAQQEQQIQ 220
E SD GA+ EQ ++
Sbjct 159 EENSDEGAEVEQIVE 173
Lambda K H
0.312 0.127 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7266557660
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40