bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_3027_orf1
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
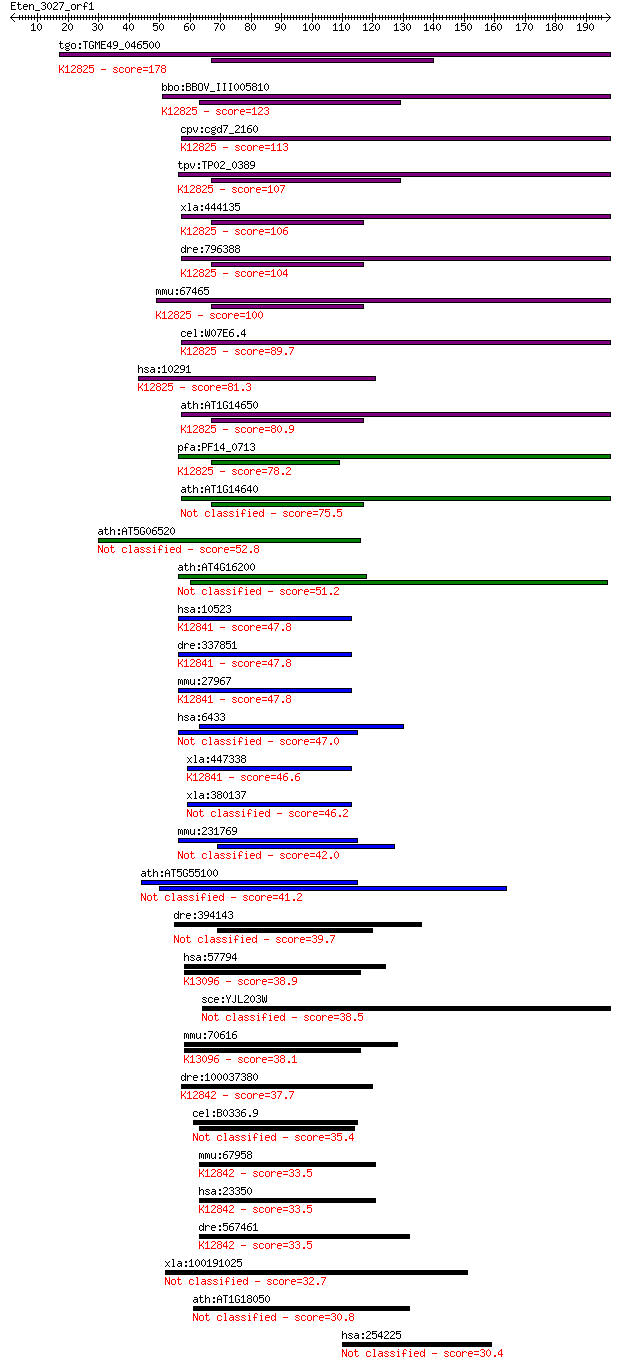
tgo:TGME49_046500 surp module domain-containing protein ; K128... 178 1e-44
bbo:BBOV_III005810 17.m07515; Surp module domain containing pr... 123 5e-28
cpv:cgd7_2160 Pre-mRNA splicing factor SF3a. 2xSWAP domain pro... 113 3e-25
tpv:TP02_0389 hypothetical protein; K12825 splicing factor 3A ... 107 2e-23
xla:444135 MGC80562 protein; K12825 splicing factor 3A subunit 1 106 4e-23
dre:796388 sf3a1, MGC65786, MGC77239, si:dz150f13.2, wu:fc38e0... 104 2e-22
mmu:67465 Sf3a1, 1200014H24Rik, 5930416L09Rik, AI159724; splic... 100 3e-21
cel:W07E6.4 prp-21; yeast PRP (splicing factor) related family... 89.7 5e-18
hsa:10291 SF3A1, PRP21, PRPF21, SAP114, SF3A120; splicing fact... 81.3 2e-15
ath:AT1G14650 SWAP (Suppressor-of-White-APricot)/surp domain-c... 80.9 3e-15
pfa:PF14_0713 conserved Plasmodium protein, unknown function; ... 78.2 2e-14
ath:AT1G14640 SWAP (Suppressor-of-White-APricot)/surp domain-c... 75.5 1e-13
ath:AT5G06520 SWAP (Suppressor-of-White-APricot)/surp domain-c... 52.8 8e-07
ath:AT4G16200 SWAP (Suppressor-of-White-APricot)/surp domain-c... 51.2 2e-06
hsa:10523 CHERP, DAN16, SCAF6, SRA1; calcium homeostasis endop... 47.8 2e-05
dre:337851 cherp, MGC55518, ik:tdsubc_2h12, scaf6, wu:fc83d01,... 47.8 2e-05
mmu:27967 Cherp, 5730408I11Rik, D8Wsu96e, DAN16, Scaf6; calciu... 47.8 3e-05
hsa:6433 SFSWAP, MGC167082, SFRS8, SWAP; splicing factor, supp... 47.0 4e-05
xla:447338 cherp, MGC83231; calcium homeostasis endoplasmic re... 46.6 5e-05
xla:380137 cherp, MGC53695; calcium homeostasis endoplasmic re... 46.2 7e-05
mmu:231769 Sfswap, 1190005N23Rik, 6330437E22Rik, AI197402, AW2... 42.0 0.001
ath:AT5G55100 SWAP (Suppressor-of-White-APricot)/surp domain-c... 41.2 0.002
dre:394143 sfswap, MGC56184, sfrs8, srsf8, zgc:56184; splicing... 39.7 0.006
hsa:57794 SUGP1, DKFZp434E2216, F23858, RBP, SF4; SURP and G p... 38.9 0.012
sce:YJL203W PRP21, SPP91; Prp21p 38.5 0.016
mmu:70616 Sugp1, 5730496N02Rik, Sf4; SURP and G patch domain c... 38.1 0.019
dre:100037380 zgc:163098; K12842 U2-associated protein SR140 37.7 0.027
cel:B0336.9 swp-1; splicing factor (Suppressor of White aPrico... 35.4 0.14
mmu:67958 2610101N10Rik, AU023006, Sr140, U2surp; RIKEN cDNA 2... 33.5 0.43
hsa:23350 U2SURP, KIAA0332, MGC133197, SR140, fSAPa; U2 snRNP-... 33.5 0.43
dre:567461 KIAA0332, mp:zf637-1-000918, wu:fa03e02, wu:fb38b01... 33.5 0.47
xla:100191025 sall2, MGC82592, Xsal2, XsalF, XsalF-a, XsalF-b,... 32.7 0.73
ath:AT1G18050 SWAP (Suppressor-of-White-APricot)/surp domain-c... 30.8 3.1
hsa:254225 RNF169, KIAA1991; ring finger protein 169 30.4
> tgo:TGME49_046500 surp module domain-containing protein ; K12825
splicing factor 3A subunit 1
Length=683
Score = 178 bits (451), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 98/181 (54%), Positives = 127/181 (70%), Gaps = 11/181 (6%)
Query 17 MSAAVEPTQRPSGSPTGPGASGASGVGTSDVGAPGTPHPDLIFPPADMRGVIEKTAQFVA 76
MSAAV P P PG S A GVG ++FPP D+RG+IEKTAQFVA
Sbjct 26 MSAAVLP------PPVAPGDSNA-GVGDPKASGSAVNPSSIVFPPPDLRGIIEKTAQFVA 78
Query 77 KNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVREFTTGEAAPTPQVPQAIRDMQQK 136
KNG EFE RV REQ+ +F FL PNNPY+A+Y+LKVREF TGE AP PQVPQAI DM++K
Sbjct 79 KNGVEFEQRVRREQNNQRFAFLFPNNPYNAYYQLKVREFQTGEEAPRPQVPQAILDMRKK 138
Query 137 QQQEKQQQQQLLMLTQIGESEKEANKEQIEPPPPHQFVLQHPWVAPIDLDIIRCAAQFVA 196
+++E+++++++LMLTQ GE K++IEPP P + + P++A ID+DII+ AQFVA
Sbjct 139 EEEERKKKERVLMLTQYGEEA----KKKIEPPAPDVYSVTQPYIALIDVDIIQTTAQFVA 194
Query 197 R 197
R
Sbjct 195 R 195
Score = 35.4 bits (80), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 39/74 (52%), Gaps = 2/74 (2%)
Query 67 VIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREFTTGEAAPTPQ 125
+I+ TAQFVA+NG F S + RE+ +F FL P + ++ V +T P+ Q
Sbjct 185 IIQTTAQFVARNGQRFLSGLAQRERQNPQFEFLKPTHALFQYFTNLVDAYTKCLLPPSEQ 244
Query 126 VPQAIRDMQQKQQQ 139
+ + I D+ Q+
Sbjct 245 M-EKISDIAGSTQK 257
> bbo:BBOV_III005810 17.m07515; Surp module domain containing
protein; K12825 splicing factor 3A subunit 1
Length=482
Score = 123 bits (308), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 66/154 (42%), Positives = 92/154 (59%), Gaps = 10/154 (6%)
Query 51 GTPHPDLIFPPADMRGVIEKTAQFVAKNGAEFESRVLREQS------ATKFGFLLPNNPY 104
G D+IFPP +R +I+KTAQFV+KNG +FE R+ EQS KF FL P+N Y
Sbjct 2 GNKIEDIIFPPKYVRTIIDKTAQFVSKNGEQFEQRLRAEQSDGAAGAGAKFAFLSPDNAY 61
Query 105 HAFYKLKVREFTTG-EAAPTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKE 163
HA+YKLK+ E G + P +PQAI D ++K + + Q +++LL LT G S +
Sbjct 62 HAYYKLKLTELRNGIDVEIKPSIPQAILDRRKKLELKNQFKERLLALTDFGSSSAD---H 118
Query 164 QIEPPPPHQFVLQHPWVAPIDLDIIRCAAQFVAR 197
++ P F P+VA +D+DII+ A FVAR
Sbjct 119 ELGEPETDTFSFTQPFVASMDMDIIKATACFVAR 152
Score = 33.1 bits (74), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Query 63 DMRGVIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREFTTGEAA 121
DM +I+ TA FVA+NG F + RE++ +F FL P++ F+ +T
Sbjct 139 DM-DIIKATACFVARNGQRFLVELTKREKNNPQFDFLNPSHCLFGFFTNLTESYTKCLLP 197
Query 122 PTPQVPQ 128
PQV +
Sbjct 198 TKPQVAK 204
> cpv:cgd7_2160 Pre-mRNA splicing factor SF3a. 2xSWAP domain protein
; K12825 splicing factor 3A subunit 1
Length=462
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 54/145 (37%), Positives = 87/145 (60%), Gaps = 4/145 (2%)
Query 57 LIFPPADMRGVIEKTAQFVAKNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVREFT 116
LI+PP ++R I+KTA FVAKNG EFESR+L E + KF FL +NP+H +YK ++ +F
Sbjct 9 LIYPPFELRATIDKTASFVAKNGEEFESRILSESGSIKFTFLNKDNPFHLYYKKRIEDFK 68
Query 117 TGEAAPT--PQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQIEPPPPH--Q 172
G + P +P+AI DM +++++ ++++LMLT +EP P Q
Sbjct 69 NGVSIDNSGPTIPRAILDMNSRKEKQIIAEKEVLMLTSFSGGFGFMGGAVMEPEEPRKDQ 128
Query 173 FVLQHPWVAPIDLDIIRCAAQFVAR 197
+ + HP ++ D +I+ A ++AR
Sbjct 129 YTISHPIISIKDESVIKITAMYLAR 153
> tpv:TP02_0389 hypothetical protein; K12825 splicing factor 3A
subunit 1
Length=443
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 57/149 (38%), Positives = 89/149 (59%), Gaps = 12/149 (8%)
Query 56 DLIFPPADMRGVIEKTAQFVAKNGAEFESRVLREQSAT------KFGFLLPNNPYHAFYK 109
D+IFPP+ +R VI+KTA+FVAKNG +F S++ +QS + KF FL P N YH +YK
Sbjct 6 DIIFPPSFIRSVIDKTAEFVAKNGEQFVSKLRLDQSNSSLNDNIKFNFLEPGNAYHLYYK 65
Query 110 LKVREFTTGEAAP-TPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQIEPP 168
LK+ E + A P +PQAI D ++K + + + +++LL L+ S E ++ P
Sbjct 66 LKLSELQGNKVADLNPSIPQAILDKREKLELKTKHKERLLALSDFRHS-----SESVQKP 120
Query 169 PPHQFVLQHPWVAPIDLDIIRCAAQFVAR 197
F P+++ +D ++I+ A FVAR
Sbjct 121 EDDLFSFTMPFISALDYEVIKNTALFVAR 149
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 1/63 (1%)
Query 67 VIEKTAQFVAKNGAEFESRV-LREQSATKFGFLLPNNPYHAFYKLKVREFTTGEAAPTPQ 125
VI+ TA FVA+NG +F + RE++ ++ FL P++ F+ +T P Q
Sbjct 139 VIKNTALFVARNGHKFLVDLNKREKNNPQYDFLNPSHYLFTFFSNLTDSYTKCLLPPRSQ 198
Query 126 VPQ 128
+ +
Sbjct 199 IDK 201
> xla:444135 MGC80562 protein; K12825 splicing factor 3A subunit
1
Length=802
Score = 106 bits (265), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 59/143 (41%), Positives = 92/143 (64%), Gaps = 12/143 (8%)
Query 57 LIFPPADMRGVIEKTAQFVAKNGAEFESRVLR-EQSATKFGFLLPNNPYHAFYKLKVREF 115
+I+PP ++R +++KTA FVA+NG EFE+R+ + E + KF FL PN+PYHA+Y+ KV EF
Sbjct 37 IIYPPPEVRNIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYHAYYRHKVNEF 96
Query 116 TTGEA-APTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQIEPPPPHQFV 174
G+A P+ +P+ ++ Q QQ ++ Q Q++ T I + EPPP ++FV
Sbjct 97 KEGKAQEPSAAIPKVMQQQQSVQQLPQKLQAQVVQETIIPK----------EPPPEYEFV 146
Query 175 LQHPWVAPIDLDIIRCAAQFVAR 197
P ++ DLD+++ AQFVAR
Sbjct 147 ADPPSISAFDLDVVKLTAQFVAR 169
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query 67 VIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREFT 116
V++ TAQFVA+NG +F ++++ +EQ +F FL P + ++ V ++T
Sbjct 159 VVKLTAQFVARNGRQFLTQLMQKEQRNYQFDFLRPQHSLFNYFTKLVEQYT 209
> dre:796388 sf3a1, MGC65786, MGC77239, si:dz150f13.2, wu:fc38e01,
wu:fc50a11, wu:fj37c05, zgc:65786; splicing factor 3a,
subunit 1; K12825 splicing factor 3A subunit 1
Length=780
Score = 104 bits (260), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 58/143 (40%), Positives = 90/143 (62%), Gaps = 15/143 (10%)
Query 57 LIFPPADMRGVIEKTAQFVAKNGAEFESRVLR-EQSATKFGFLLPNNPYHAFYKLKVREF 115
+I+PP ++R +++KTA FVA+NG EFE+R+ + E + KF FL P++PYHA+Y+ KV EF
Sbjct 33 IIYPPPEVRNIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPSDPYHAYYRHKVNEF 92
Query 116 TTGEA-APTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQIEPPPPHQFV 174
G+A P+ VP+ MQ QQ ++ Q Q++ T + + EPPP +F+
Sbjct 93 KEGKAQEPSAAVPKV---MQHPQQLPQKVQAQVIHETVVPK----------EPPPEFEFI 139
Query 175 LQHPWVAPIDLDIIRCAAQFVAR 197
P ++ DLD+++ AQFVAR
Sbjct 140 ADPPSISAFDLDVVKLTAQFVAR 162
Score = 38.5 bits (88), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query 67 VIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREFT 116
V++ TAQFVA+NG +F ++++ +EQ +F FL P + ++ V ++T
Sbjct 152 VVKLTAQFVARNGRQFLTQLMQKEQRNYQFDFLRPQHSLFNYFTKLVEQYT 202
> mmu:67465 Sf3a1, 1200014H24Rik, 5930416L09Rik, AI159724; splicing
factor 3a, subunit 1; K12825 splicing factor 3A subunit
1
Length=791
Score = 100 bits (249), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 58/153 (37%), Positives = 93/153 (60%), Gaps = 14/153 (9%)
Query 49 APGTPHPDLIFPPADMRGVIEKTAQFVAKNGAEFESRVLR-EQSATKFGFLLPNNPYHAF 107
P P +I+PP ++R +++KTA FVA+NG EFE+R+ + E + KF FL PN+PYHA+
Sbjct 34 TPSKPVVGIIYPPPEVRNIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPNDPYHAY 93
Query 108 YKLKVREFTTGEA-APTPQVPQAI--RDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQ 164
Y+ KV EF G+A P+ +P+ + + +QQ ++ Q Q++ T + +
Sbjct 94 YRHKVSEFKEGKAQEPSAAIPKVMQQQQQATQQQLPQKVQAQVIQETIVPK--------- 144
Query 165 IEPPPPHQFVLQHPWVAPIDLDIIRCAAQFVAR 197
EPPP +F+ P ++ DLD+++ AQFVAR
Sbjct 145 -EPPPEFEFIADPPSISAFDLDVVKLTAQFVAR 176
Score = 38.5 bits (88), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 33/51 (64%), Gaps = 1/51 (1%)
Query 67 VIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREFT 116
V++ TAQFVA+NG +F ++++ +EQ +F FL P + ++ V ++T
Sbjct 166 VVKLTAQFVARNGRQFLTQLMQKEQRNYQFDFLRPQHSLFNYFTKLVEQYT 216
> cel:W07E6.4 prp-21; yeast PRP (splicing factor) related family
member (prp-21); K12825 splicing factor 3A subunit 1
Length=655
Score = 89.7 bits (221), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 54/143 (37%), Positives = 77/143 (53%), Gaps = 27/143 (18%)
Query 57 LIFPPADMRGVIEKTAQFVAKNGAEFESRVLREQSAT--KFGFLLPNNPYHAFYKLKVRE 114
LI+PP D+R +++KTA+F AKNG +FE+++ RE+ A KF FL +PYHA+YK V +
Sbjct 27 LIYPPPDIRTIVDKTARFAAKNGVDFENKI-REKEAKNPKFNFLSITDPYHAYYKKMVYD 85
Query 115 FTTGEAAPTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQIEPPPPHQFV 174
F+ G P+VPQA++ E K+A PPP ++F
Sbjct 86 FSEGRVE-APKVPQAVK-----------------------EHVKKAEFVPSAPPPAYEFS 121
Query 175 LQHPWVAPIDLDIIRCAAQFVAR 197
+ DLD+IR A FVAR
Sbjct 122 ADPSTINAYDLDLIRLVALFVAR 144
> hsa:10291 SF3A1, PRP21, PRPF21, SAP114, SF3A120; splicing factor
3a, subunit 1, 120kDa; K12825 splicing factor 3A subunit
1
Length=728
Score = 81.3 bits (199), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/79 (46%), Positives = 55/79 (69%), Gaps = 1/79 (1%)
Query 43 GTSDVGAPGTPHPDLIFPPADMRGVIEKTAQFVAKNGAEFESRVLR-EQSATKFGFLLPN 101
+ + AP P +I+PP ++R +++KTA FVA+NG EFE+R+ + E + KF FL PN
Sbjct 28 SSKEDSAPSKPVVGIIYPPPEVRNIVDKTASFVARNGPEFEARIRQNEINNPKFNFLNPN 87
Query 102 NPYHAFYKLKVREFTTGEA 120
+PYHA+Y+ KV EF G+A
Sbjct 88 DPYHAYYRHKVSEFKEGKA 106
> ath:AT1G14650 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein / ubiquitin family protein; K12825 splicing
factor 3A subunit 1
Length=785
Score = 80.9 bits (198), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 50/154 (32%), Positives = 78/154 (50%), Gaps = 24/154 (15%)
Query 57 LIFPPADMRGVIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREF 115
+I PP D+R ++EKTAQFV+KNG EFE R++ + KF FL ++PYHAFY+ K+ E+
Sbjct 61 IIHPPPDIRTIVEKTAQFVSKNGLEFEKRIIVSNEKNAKFNFLKSSDPYHAFYQHKLTEY 120
Query 116 -----------TTGEAAPTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQ 164
+ PQ+ D + + Q Q + ++
Sbjct 121 RAQNKDGAQGTDDSDGTTDPQLDTGAADESEAGDTQPDLQAQFRIPSK-----------P 169
Query 165 IEPPPPHQFVLQHP-WVAPIDLDIIRCAAQFVAR 197
+E P P ++ ++ P + +LDII+ AQFVAR
Sbjct 170 LEAPEPEKYTVRLPEGITGEELDIIKLTAQFVAR 203
Score = 33.1 bits (74), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 67 VIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREFT 116
+I+ TAQFVA+NG F + + RE + +F F+ P + F+ V ++
Sbjct 193 IIKLTAQFVARNGKSFLTGLSNRENNNPQFHFMKPTHSMFTFFTSLVDAYS 243
> pfa:PF14_0713 conserved Plasmodium protein, unknown function;
K12825 splicing factor 3A subunit 1
Length=701
Score = 78.2 bits (191), Expect = 2e-14, Method: Composition-based stats.
Identities = 46/163 (28%), Positives = 83/163 (50%), Gaps = 22/163 (13%)
Query 56 DLIFPPADMRGVIEKTAQFVAKNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVREF 115
++I PP +++ VI+KTA FV KNG FE ++ RE+ +FGF+ P++PY +Y+ K+
Sbjct 33 EMIVPPNNIKTVIDKTATFVKKNGKNFEQKIYREKE-KQFGFISPSHPYFYYYQYKLHGL 91
Query 116 TTGEAAPTPQVPQAIRDMQQKQQQEKQQQQQ--LLMLTQIGESEKEANK----------- 162
+P I+++++++ K + L + I E EKE K
Sbjct 92 FLDIQEKDELIPHVIKEIKKREDANKYTNNEHILKICDFIKEDEKEERKNIIYSLDDMKK 151
Query 163 -EQIEPPPPHQFVLQH-------PWVAPIDLDIIRCAAQFVAR 197
+++E + +Q P++ +D+D+I+ A FVAR
Sbjct 152 IDKLENNNNLKIQIQEDIYSLISPFITSLDIDLIKTTALFVAR 194
Score = 34.3 bits (77), Expect = 0.30, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 29/43 (67%), Gaps = 1/43 (2%)
Query 67 VIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFY 108
+I+ TA FVA+NG F + ++ RE++ +++ FL NN Y ++
Sbjct 184 LIKTTALFVARNGKSFLNGLIEREKNNSQYDFLRANNLYFNYF 226
> ath:AT1G14640 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=735
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 47/146 (32%), Positives = 80/146 (54%), Gaps = 16/146 (10%)
Query 57 LIFPPADMRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYHAFYKLKVREF 115
+I+PP ++R ++E TAQFV++NG F ++V E++ F FL +NPYH FY+ KV E+
Sbjct 60 IIYPPPEIRKIVETTAQFVSQNGLAFGNKVKTEKANNANFSFLKSDNPYHGFYRYKVTEY 119
Query 116 TTGEAAPTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIG---ESEKEANKEQIEPPPPHQ 172
+ IRD Q + + +L + +++ A ++ +E P P +
Sbjct 120 SC-----------HIRDGAQGTDVDDTEDPKLDDESDAKPDLQAQFRAPRKILEAPEPEK 168
Query 173 FVLQHP-WVAPIDLDIIRCAAQFVAR 197
+ ++ P + +LDII+ AQFVAR
Sbjct 169 YTVRLPEGIMEAELDIIKHTAQFVAR 194
Score = 35.8 bits (81), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query 67 VIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNPYHAFYKLKVREFT 116
+I+ TAQFVA+NG F ++ RE + ++F F+ P + F+ V ++
Sbjct 184 IIKHTAQFVARNGQSFLRELMRREVNNSQFQFMKPTHSMFTFFTSLVDAYS 234
> ath:AT5G06520 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=679
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 48/87 (55%), Gaps = 4/87 (4%)
Query 30 SPTGPGASGASGVGTSDVGAPGTPHPDLIFPPADMRGVIEKTAQFVAKNGAEFESRVLR- 88
SP P SG S+ AP + P I PP ++R +E TA V+KNG E E +++
Sbjct 370 SPPRPQNDLQSGQSNSN-KAPASVAP--IEPPPEIRSCVENTALIVSKNGLEIERKMMEL 426
Query 89 EQSATKFGFLLPNNPYHAFYKLKVREF 115
+ + F+ +PYHAFY+LK+ E+
Sbjct 427 SMNDARHRFVWSTDPYHAFYQLKLAEY 453
> ath:AT4G16200 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=288
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 44/66 (66%), Gaps = 4/66 (6%)
Query 56 DLIFPPADM--RGVIEKTAQFVAKNGAEFESRVLREQ-SATKFGFLLPN-NPYHAFYKLK 111
++I PPAD+ R +++K AQFV+K G EFE++++ + KF FL +P H +YK K
Sbjct 13 EIITPPADIGTRTLVDKAAQFVSKKGLEFETKIIDSYPTDAKFNFLRSTADPCHTYYKHK 72
Query 112 VREFTT 117
+ E+++
Sbjct 73 LAEYSS 78
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 60/142 (42%), Gaps = 31/142 (21%)
Query 60 PPADMRGVIEKTAQFVAKNGAEFESRVLREQS-ATKFGFLLPN-NPYHAFYKLKVREFTT 117
P ++E T+ V++ G+EFE V + +F FL + +PY+ YK K+ E++
Sbjct 97 PNVTTTAIVETTSCLVSQFGSEFEMMVKDSNTDDARFNFLKSSEDPYNVLYKQKLDEYSL 156
Query 118 GEAAPTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQIEPPPPHQFVLQH 177
D + Q K + I + +EA + + P +LQ
Sbjct 157 --------------DPRDAYYQHK-------LAESISKYHREATPKLVLPN-----LLQF 190
Query 178 PWVAPI---DLDIIRCAAQFVA 196
V + +LD ++ AQFVA
Sbjct 191 RLVKEMTFEELDTVKVTAQFVA 212
> hsa:10523 CHERP, DAN16, SCAF6, SRA1; calcium homeostasis endoplasmic
reticulum protein; K12841 calcium homeostasis endoplasmic
reticulum protein
Length=916
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 37/60 (61%), Gaps = 4/60 (6%)
Query 56 DLIFPPAD--MRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYHAFYKLKV 112
++ PP D +R VI+K AQFVA+NG EFE + +Q KF FL ++++YK K+
Sbjct 2 EMPLPPDDQELRNVIDKLAQFVARNGPEFEKMTMEKQKDNPKFSFLF-GGEFYSYYKCKL 60
> dre:337851 cherp, MGC55518, ik:tdsubc_2h12, scaf6, wu:fc83d01,
xx:tdsubc_2h12, zgc:55518; calcium homeostasis endoplasmic
reticulum protein; K12841 calcium homeostasis endoplasmic
reticulum protein
Length=909
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/60 (43%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Query 56 DLIFPPAD--MRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYHAFYKLKV 112
D+ PP D +R VI+K AQFVA+NG EFE + +Q KF FL + +YK K+
Sbjct 2 DISTPPEDQELRNVIDKLAQFVARNGPEFEKMTMEKQKDNAKFSFLF-GGEFFGYYKYKL 60
> mmu:27967 Cherp, 5730408I11Rik, D8Wsu96e, DAN16, Scaf6; calcium
homeostasis endoplasmic reticulum protein; K12841 calcium
homeostasis endoplasmic reticulum protein
Length=938
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 37/60 (61%), Gaps = 4/60 (6%)
Query 56 DLIFPPAD--MRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYHAFYKLKV 112
++ PP D +R VI+K AQFVA+NG EFE + +Q KF FL ++++YK K+
Sbjct 2 EMPMPPDDQELRNVIDKLAQFVARNGPEFEKMTMEKQKDNPKFSFLF-GGEFYSYYKCKL 60
> hsa:6433 SFSWAP, MGC167082, SFRS8, SWAP; splicing factor, suppressor
of white-apricot homolog (Drosophila)
Length=951
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 63 DMRGVIEKTAQFVAKNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVREFTTGEAAP 122
D++ VI+K A++VA+NG +FE+ V R ++ +F FL P + Y+A+Y+ K + F E
Sbjct 455 DVQPVIDKLAEYVARNGLKFETSV-RAKNDQRFEFLQPWHQYNAYYEFKKQFFLQKEGGD 513
Query 123 TPQVPQA 129
+ Q A
Sbjct 514 SMQAVSA 520
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 56 DLIFPP-ADMRGVIEKTAQFVAKNGAEFESRVLREQS-ATKFGFLLPN---NPYHAFYKL 110
D+ PP A M +IE+TA FV + GA+FE + +Q+ ++F FL + NPY+ F +
Sbjct 199 DVELPPTAKMHAIIERTASFVCRQGAQFEIMLKAKQARNSQFDFLRFDHYLNPYYKFIQK 258
Query 111 KVRE 114
++E
Sbjct 259 AMKE 262
> xla:447338 cherp, MGC83231; calcium homeostasis endoplasmic
reticulum protein; K12841 calcium homeostasis endoplasmic reticulum
protein
Length=933
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 34/57 (59%), Gaps = 4/57 (7%)
Query 59 FPPAD--MRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYHAFYKLKV 112
PP D +R VI+K AQFVA+NG EFE + +Q KF FL ++ +YK K+
Sbjct 5 LPPEDQELRNVIDKLAQFVARNGPEFEKMTMEKQKDNHKFSFLF-GGEFYNYYKCKL 60
> xla:380137 cherp, MGC53695; calcium homeostasis endoplasmic
reticulum protein
Length=924
Score = 46.2 bits (108), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 33/57 (57%), Gaps = 4/57 (7%)
Query 59 FPPAD--MRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYHAFYKLKV 112
PP D +R VI+K AQFVA+NG EFE + +Q KF FL Y +Y+ K+
Sbjct 5 LPPEDQELRNVIDKLAQFVARNGPEFEKMTMEKQKDNLKFSFLF-GGEYFNYYRCKL 60
> mmu:231769 Sfswap, 1190005N23Rik, 6330437E22Rik, AI197402, AW212079,
SWAP, Sfrs8, Srsf8; splicing factor, suppressor of
white-apricot homolog (Drosophila)
Length=945
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 39/64 (60%), Gaps = 5/64 (7%)
Query 56 DLIFPP-ADMRGVIEKTAQFVAKNGAEFESRVLREQS-ATKFGFLLPN---NPYHAFYKL 110
D+ PP A M +IE+TA FV K GA+FE + +Q+ ++F FL + NPY+ F +
Sbjct 199 DVELPPTAKMHAIIERTANFVCKQGAQFEIMLKAKQARNSQFDFLRFDHYLNPYYKFIQK 258
Query 111 KVRE 114
++E
Sbjct 259 AMKE 262
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 36/58 (62%), Gaps = 1/58 (1%)
Query 69 EKTAQFVAKNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVREFTTGEAAPTPQV 126
+K A++VA+NG +FE+ V R ++ +F FL P + Y+A+Y+ K + F E + Q
Sbjct 460 DKLAEYVARNGLKFETSV-RAKNDQRFEFLQPWHQYNAYYEFKKQFFLQKEGGGSTQA 516
> ath:AT5G55100 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=844
Score = 41.2 bits (95), Expect = 0.002, Method: Composition-based stats.
Identities = 25/72 (34%), Positives = 40/72 (55%), Gaps = 2/72 (2%)
Query 44 TSDVGAPGTPHPDLIFPPADMRGVIEKTAQFVAKNGAEFESR-VLREQSATKFGFLLPNN 102
TSD P ++ P +M+ VI+K F+ KNG E E+ V ++ F FL P++
Sbjct 316 TSDKSQPKL-ELQIVEPTTEMKRVIDKIVDFIQKNGKELEATLVAQDVKYGMFPFLRPSS 374
Query 103 PYHAFYKLKVRE 114
YHA+Y+ ++E
Sbjct 375 LYHAYYRKVLQE 386
Score = 38.1 bits (87), Expect = 0.021, Method: Composition-based stats.
Identities = 36/123 (29%), Positives = 62/123 (50%), Gaps = 20/123 (16%)
Query 50 PGTPHPDLI---FPPAD-MRGVIEKTAQFVAKNGAEFESRVLREQSATK--FGFLLPNNP 103
P P PD + PP + + +I +T+ FV+K+G + E VLR + FGFL+P++
Sbjct 132 PPFPVPDYLRQNLPPTEKLHQIITRTSSFVSKHGGQSEI-VLRVKQGDNPTFGFLMPDHH 190
Query 104 YHAFYKLKV--REFTTGEAAPTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKE-A 160
H +++ V +E TG+++ ++K + EK L+ + G E E A
Sbjct 191 LHLYFRFLVDHQELLTGKSSVE----------EKKNESEKDGGALSLLGSVYGTVEDEDA 240
Query 161 NKE 163
N+E
Sbjct 241 NEE 243
> dre:394143 sfswap, MGC56184, sfrs8, srsf8, zgc:56184; splicing
factor, suppressor of white-apricot homolog (Drosophila)
Length=958
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 13/90 (14%)
Query 55 PDLIFPP-------ADMRGVIEKTAQFVAKNGAEFESRVLREQSA--TKFGFLLPNNPYH 105
P L+ PP +IE+TA FV K GA+FE VL+ + A ++F FL ++ +
Sbjct 180 PGLLIPPDVELPATTKTHAIIERTANFVCKQGAQFEI-VLKAKQAGNSQFDFLRFDHYLN 238
Query 106 AFYKLKVREFTTGEAAPTPQVPQAIRDMQQ 135
+YK +R G P + Q D QQ
Sbjct 239 PYYKHILRAMKEGRYTPASEGKQ---DQQQ 265
Score = 38.1 bits (87), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query 69 EKTAQFVAKNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVREFTTGE 119
+K A++VA+NG +FE+ V R ++ +F FL + Y+++Y+ K F E
Sbjct 480 DKLAEYVARNGVKFETSV-RAKNDPRFDFLQSWHQYNSYYEFKKHYFMQKE 529
> hsa:57794 SUGP1, DKFZp434E2216, F23858, RBP, SF4; SURP and G
patch domain containing 1; K13096 splicing factor 4
Length=645
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 39/70 (55%), Gaps = 4/70 (5%)
Query 58 IFPPAD--MRGVIEKTAQFVAKNGAEFESRVLREQSATK-FGFLL-PNNPYHAFYKLKVR 113
+ PP D ++ + EK A+F+A G E E+ L+ + F FL PN+ + +Y+ K+
Sbjct 255 VSPPEDEEVKNLAEKLARFIADGGPEVETIALQNNRENQAFSFLYEPNSQGYKYYRQKLE 314
Query 114 EFTTGEAAPT 123
EF +A+ T
Sbjct 315 EFRKAKASST 324
Score = 33.1 bits (74), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 4/62 (6%)
Query 58 IFPP--ADMRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYH-AFYKLKVR 113
+ PP A+ R VIEK A+FVA+ G E E + + F FL N +Y+ KV
Sbjct 180 VSPPEGAETRKVIEKLARFVAEGGPELEKVAMEDYKDNPAFAFLHDKNSREFLYYRKKVA 239
Query 114 EF 115
E
Sbjct 240 EI 241
> sce:YJL203W PRP21, SPP91; Prp21p
Length=280
Score = 38.5 bits (88), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 26/134 (19%), Positives = 56/134 (41%), Gaps = 36/134 (26%)
Query 64 MRGVIEKTAQFVAKNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVREFTTGEAAPT 123
++ I+ T ++ ++G EFE+++L ++ +F F+ ++P H +Y + E PT
Sbjct 8 LKEDIKTTVNYIKQHGVEFENKLLEDE---RFSFIKKDDPLHEYYTKLMNE-------PT 57
Query 124 PQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEKEANKEQIEPPPPHQFVLQHPWVAPI 183
V GE ++ +I PP F ++
Sbjct 58 DTVS--------------------------GEDNDRKSEREIARPPDFLFSQYDTGISRR 91
Query 184 DLDIIRCAAQFVAR 197
D+++I+ A++ A+
Sbjct 92 DMEVIKLTARYYAK 105
> mmu:70616 Sugp1, 5730496N02Rik, Sf4; SURP and G patch domain
containing 1; K13096 splicing factor 4
Length=643
Score = 38.1 bits (87), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 38/74 (51%), Gaps = 4/74 (5%)
Query 58 IFPPAD--MRGVIEKTAQFVAKNGAEFESRVLREQSATK-FGFLL-PNNPYHAFYKLKVR 113
+ PP D + + EK A+F+A G E E+ L+ + F FL PN+ + +Y+ K+
Sbjct 251 VSPPEDEEAKNLAEKLARFIADGGPEVETIALQNNRENQAFSFLYDPNSQGYRYYRQKLD 310
Query 114 EFTTGEAAPTPQVP 127
EF +A T P
Sbjct 311 EFRKAKAGSTGSFP 324
Score = 32.3 bits (72), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 31/62 (50%), Gaps = 4/62 (6%)
Query 58 IFPP--ADMRGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYH-AFYKLKVR 113
+ PP A+ R VIEK A+FVA+ G E E + + F FL N +Y+ KV
Sbjct 176 VSPPEGAETRRVIEKLARFVAEGGPELEKVAMEDYKDNPAFTFLHDKNSREFLYYRRKVA 235
Query 114 EF 115
E
Sbjct 236 EI 237
> dre:100037380 zgc:163098; K12842 U2-associated protein SR140
Length=874
Score = 37.7 bits (86), Expect = 0.027, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Query 57 LIFPPADMRGVIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPNNP-YHAFYKLKVRE 114
+I P ++ G+I + +FV + G FE+ ++ RE++ F FL N H +Y+ K+
Sbjct 310 VIPPERNLLGLIHRMIEFVVREGPMFEAMIMNREKNNPDFRFLFDNKSQEHVYYRWKLYS 369
Query 115 FTTGE 119
GE
Sbjct 370 ILQGE 374
> cel:B0336.9 swp-1; splicing factor (Suppressor of White aPricot)
related family member (swp-1)
Length=751
Score = 35.4 bits (80), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 37/61 (60%), Gaps = 7/61 (11%)
Query 61 PADMR--GVIEKTAQFVAKNGAEFESRVLREQ--SATKFGFLLPN---NPYHAFYKLKVR 113
P++M+ +IEKTA F+ NG + E + +Q +A +FGFL + NP++ + + +R
Sbjct 162 PSNMKLHHIIEKTASFIVANGTQMEIVIKAKQRNNAEQFGFLEFDHRLNPFYKYLQKLIR 221
Query 114 E 114
E
Sbjct 222 E 222
Score = 32.7 bits (73), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 63 DMRGVIEKTAQFVAKNGAEFESRVLREQSATKFGFLLPNNPYHAFYKLKVR 113
D+ ++ A+ VA+ G E E+ L + + F+ P +PY+++Y KVR
Sbjct 391 DLEPILNSYAEHVAQRGLEAEA-SLAAREDLQLHFMEPKSPYYSYYHHKVR 440
> mmu:67958 2610101N10Rik, AU023006, Sr140, U2surp; RIKEN cDNA
2610101N10 gene; K12842 U2-associated protein SR140
Length=1029
Score = 33.5 bits (75), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 63 DMRGVIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPN-NPYHAFYKLKVREFTTGEA 120
++ +I + +FV + G FE+ ++ RE + F FL N P H +Y+ K+ G++
Sbjct 426 NLLALIHRMIEFVVREGPMFEAMIMNREINNPMFRFLFENQTPAHVYYRWKLYSILQGDS 485
> hsa:23350 U2SURP, KIAA0332, MGC133197, SR140, fSAPa; U2 snRNP-associated
SURP domain containing; K12842 U2-associated protein
SR140
Length=1029
Score = 33.5 bits (75), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 63 DMRGVIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPN-NPYHAFYKLKVREFTTGEA 120
++ +I + +FV + G FE+ ++ RE + F FL N P H +Y+ K+ G++
Sbjct 426 NLLALIHRMIEFVVREGPMFEAMIMNREINNPMFRFLFENQTPAHVYYRWKLYSILQGDS 485
> dre:567461 KIAA0332, mp:zf637-1-000918, wu:fa03e02, wu:fb38b01,
wu:fc75d03; si:dkey-181m9.9; K12842 U2-associated protein
SR140
Length=968
Score = 33.5 bits (75), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 37/71 (52%), Gaps = 3/71 (4%)
Query 63 DMRGVIEKTAQFVAKNGAEFESRVL-REQSATKFGFLLPN-NPYHAFYKLKVREFTTGEA 120
++ +I + +FV + G FE+ ++ RE + + FL N +P H +Y+ K+ GE
Sbjct 360 NLLSLIHRMIEFVVREGPMFEAMIMNREINNPLYRFLFENQSPAHVYYRWKLYTILQGE- 418
Query 121 APTPQVPQAIR 131
+PT + R
Sbjct 419 SPTKWKTEDFR 429
> xla:100191025 sall2, MGC82592, Xsal2, XsalF, XsalF-a, XsalF-b,
salf; sal-like 2
Length=1106
Score = 32.7 bits (73), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 45/116 (38%), Gaps = 28/116 (24%)
Query 52 TPHPDLIFPPADMRGVIEKTAQFVAKNGAEFESRVLREQSATKFGFLLPN---------N 102
T H L+ D GVI TAQ + KN GFL PN
Sbjct 95 TAHKTLVTEQHDSTGVIGATAQDLGKNPVGLSQ-----------GFLFPNGSLIMDGLSG 143
Query 103 PYHAFYKLKVREFTTGEAAPT----PQVPQAIRDMQQKQ----QQEKQQQQQLLML 150
P + G+AA T P + + +R +QQ+Q Q +Q QQ+LML
Sbjct 144 PKFGLAPMNQETVAGGQAAATPINIPMILEELRVLQQRQIHQMQITEQICQQVLML 199
> ath:AT1G18050 SWAP (Suppressor-of-White-APricot)/surp domain-containing
protein
Length=285
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 37/83 (44%), Gaps = 12/83 (14%)
Query 61 PADM----RGVIEKTAQFVAKNGAEFESRVLREQSAT-KFGFLLPNNPYHAFYKLKVREF 115
PAD+ R + +A+ V G E E +++ + K+ F ++ YHA+Y+ K+ +
Sbjct 143 PADVPPRTRYYADSSARLVFMEGPEMERKMMTSYAGNPKYSFFWSSDRYHAYYQKKLAGY 202
Query 116 TTGEA-------APTPQVPQAIR 131
E P VP +R
Sbjct 203 QLHEGPEIKLSDYGIPDVPLRVR 225
> hsa:254225 RNF169, KIAA1991; ring finger protein 169
Length=708
Score = 30.4 bits (67), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 30/49 (61%), Gaps = 1/49 (2%)
Query 110 LKVREFTTGEAAPTPQVPQAIRDMQQKQQQEKQQQQQLLMLTQIGESEK 158
LK T+GE P P +R+M+QK QQE++ +Q L L ++ ++E+
Sbjct 637 LKKLRQTSGEVGLAPTDP-VLREMEQKLQQEEEDRQLALQLQRMFDNER 684
Lambda K H
0.316 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5802328440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40