bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2958_orf1
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
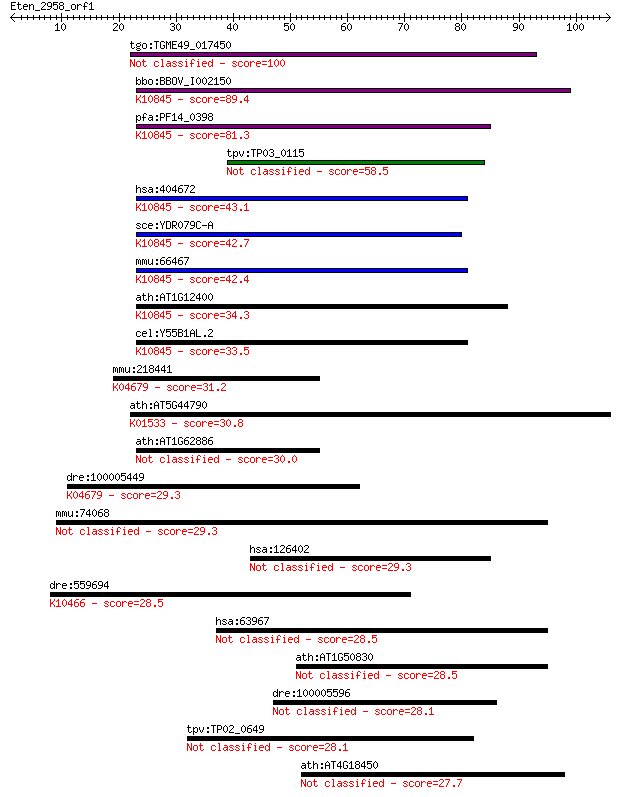
tgo:TGME49_017450 REX1 DNA repair domain-containing protein 100 2e-21
bbo:BBOV_I002150 19.m02381; hypothetical protein; K10845 TFIIH... 89.4 2e-18
pfa:PF14_0398 Transcription factor TFIIH complex subunit Tfb5,... 81.3 8e-16
tpv:TP03_0115 hypothetical protein 58.5 5e-09
hsa:404672 GTF2H5, C6orf175, TFB5, TFIIH, TGF2H5, TTD, TTD-A, ... 43.1 2e-04
sce:YDR079C-A TFB5; Component of the RNA polymerase II general... 42.7 3e-04
mmu:66467 Gtf2h5, 2700017P07Rik, 2810432H05Rik, D17Wsu155e; ge... 42.4 3e-04
ath:AT1G12400 DNA binding; K10845 TFIIH basal transcription fa... 34.3 0.097
cel:Y55B1AL.2 hypothetical protein; K10845 TFIIH basal transcr... 33.5 0.16
mmu:218441 Zfyve16, AI035632, B130024H06Rik, B130031L15, mKIAA... 31.2 0.93
ath:AT5G44790 RAN1; RAN1 (RESPONSIVE-TO-ANTAGONIST 1); ATPase,... 30.8 1.1
ath:AT1G62886 DNA binding 30.0 2.0
dre:100005449 zfyve9, si:ch211-57g18.2; zinc finger, FYVE doma... 29.3 2.9
mmu:74068 Asz1, 4933400N19Rik, Gasz, ORF3; ankyrin repeat, SAM... 29.3 3.0
hsa:126402 CCDC105, FLJ40365; coiled-coil domain containing 105 29.3
dre:559694 ectodermal-neural cortex (with BTB-like domain)-lik... 28.5 4.9
hsa:63967 CLSPN, MGC131612, MGC131613, MGC131615; claspin 28.5 5.0
ath:AT1G50830 hypothetical protein 28.5 5.4
dre:100005596 hypothetical LOC100005596 28.1 6.6
tpv:TP02_0649 hypothetical protein 28.1 6.9
ath:AT4G18450 ethylene-responsive factor, putative 27.7 8.2
> tgo:TGME49_017450 REX1 DNA repair domain-containing protein
Length=198
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 48/71 (67%), Positives = 60/71 (84%), Gaps = 0/71 (0%)
Query 22 KMVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESVCSFLEEEVTRRLEL 81
MV A+KG+LV+CDPPTME+I LNET++FIIE+++EE+VLC E V FLEEEVT+RLEL
Sbjct 112 NMVAALKGVLVKCDPPTMEVIRLLNETRDFIIEQLNEEYVLCRECVFDFLEEEVTKRLEL 171
Query 82 AERPLAEMQKE 92
AER AE Q+E
Sbjct 172 AERQTAEQQRE 182
> bbo:BBOV_I002150 19.m02381; hypothetical protein; K10845 TFIIH
basal transcription factor complex TTD-A subunit
Length=75
Score = 89.4 bits (220), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/76 (57%), Positives = 57/76 (75%), Gaps = 1/76 (1%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESVCSFLEEEVTRRLELA 82
MV +++GILV+CDPPTMEI+ Q+NET++FIIE++ E LC ESV FL EEVTRRLE A
Sbjct 1 MVTSMRGILVKCDPPTMEIVKQINETRHFIIEQLDESVALCQESVFDFLSEEVTRRLEFA 60
Query 83 ERPLAEMQKEKKANEA 98
ER L + A+E+
Sbjct 61 ER-LTPTDAKTNADES 75
> pfa:PF14_0398 Transcription factor TFIIH complex subunit Tfb5,
putative; K10845 TFIIH basal transcription factor complex
TTD-A subunit
Length=67
Score = 81.3 bits (199), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 40/62 (64%), Positives = 50/62 (80%), Gaps = 0/62 (0%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESVCSFLEEEVTRRLELA 82
MV AIKG+LV+CD PTM+IIL LNE KNF+IEKIS+ LC E+V FLE+EV ++LE +
Sbjct 1 MVTAIKGVLVKCDEPTMQIILMLNEEKNFLIEKISDTVCLCKENVHDFLEKEVIKQLEYS 60
Query 83 ER 84
ER
Sbjct 61 ER 62
> tpv:TP03_0115 hypothetical protein
Length=54
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 37/45 (82%), Gaps = 0/45 (0%)
Query 39 MEIILQLNETKNFIIEKISEEHVLCSESVCSFLEEEVTRRLELAE 83
MEI+ Q+NE+K+FIIE+I E LC +SV FL+EEVTRRLE+AE
Sbjct 1 MEIVKQINESKHFIIEQIDEGVALCKDSVTQFLKEEVTRRLEIAE 45
> hsa:404672 GTF2H5, C6orf175, TFB5, TFIIH, TGF2H5, TTD, TTD-A,
TTDA, bA120J8.2; general transcription factor IIH, polypeptide
5; K10845 TFIIH basal transcription factor complex TTD-A
subunit
Length=71
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 4/62 (6%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNET----KNFIIEKISEEHVLCSESVCSFLEEEVTRR 78
MV +KG+L++CDP + +L L+E+ K FII+ I + HV + + L+E V
Sbjct 1 MVNVLKGVLIECDPAMKQFLLYLDESNALGKKFIIQDIDDTHVFVIAELVNVLQERVGEL 60
Query 79 LE 80
++
Sbjct 61 MD 62
> sce:YDR079C-A TFB5; Component of the RNA polymerase II general
transcription and DNA repair factor TFIIH; involved in transcription
initiation and in nucleotide-excision repair; homolog
of Chlamydomonas reinhardtii REX1-S protein involved in
DNA repair; K10845 TFIIH basal transcription factor complex
TTD-A subunit
Length=72
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 36/58 (62%), Gaps = 1/58 (1%)
Query 23 MVVAIKGILVQCDPPTMEIILQLN-ETKNFIIEKISEEHVLCSESVCSFLEEEVTRRL 79
M A KG LVQCDP +ILQ++ + + ++E++ + H+L + S F++ E+ R L
Sbjct 1 MARARKGALVQCDPSIKALILQIDAKMSDIVLEELDDTHLLVNPSKVEFVKHELNRLL 58
> mmu:66467 Gtf2h5, 2700017P07Rik, 2810432H05Rik, D17Wsu155e;
general transcription factor IIH, polypeptide 5; K10845 TFIIH
basal transcription factor complex TTD-A subunit
Length=71
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 35/62 (56%), Gaps = 4/62 (6%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNET----KNFIIEKISEEHVLCSESVCSFLEEEVTRR 78
MV +KG+L++CDP + +L L+E K FII+ I + HV + + L+E V
Sbjct 1 MVNVLKGVLIECDPAMKQFLLYLDEANALGKKFIIQDIDDTHVFVIAELVNVLQERVGEL 60
Query 79 LE 80
++
Sbjct 61 MD 62
> ath:AT1G12400 DNA binding; K10845 TFIIH basal transcription
factor complex TTD-A subunit
Length=71
Score = 34.3 bits (77), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 33/71 (46%), Gaps = 6/71 (8%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNE----TKNFIIEKISEEHVLCSESVCSFLEEEVT-- 76
MV AIKG+ V CD P + I+ +N ++ FII + H+ V + ++
Sbjct 1 MVNAIKGVFVSCDIPMTQFIVNMNNSMPPSQKFIIHVLDSTHLFVQPHVEQMIRSAISDF 60
Query 77 RRLELAERPLA 87
R E+P +
Sbjct 61 RDQNSYEKPTS 71
> cel:Y55B1AL.2 hypothetical protein; K10845 TFIIH basal transcription
factor complex TTD-A subunit
Length=71
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 35/62 (56%), Gaps = 4/62 (6%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNETKN----FIIEKISEEHVLCSESVCSFLEEEVTRR 78
MV KG+LV DP ++++ L++++ FI+ ++ + H+ + + LE +V +
Sbjct 1 MVNVKKGVLVTSDPAFRQLLIHLDDSRQLGSKFIVRELDDTHLFIEKEIVPMLENKVEQI 60
Query 79 LE 80
+E
Sbjct 61 ME 62
> mmu:218441 Zfyve16, AI035632, B130024H06Rik, B130031L15, mKIAA0305;
zinc finger, FYVE domain containing 16; K04679 MAD,
mothers against decapentaplegic interacting protein
Length=1528
Score = 31.2 bits (69), Expect = 0.93, Method: Composition-based stats.
Identities = 17/37 (45%), Positives = 23/37 (62%), Gaps = 1/37 (2%)
Query 19 FLAKMVVAIKGILVQCDPPTME-IILQLNETKNFIIE 54
FLAK + G++VQ P TME + L L E K+F I+
Sbjct 1321 FLAKSSIVEDGLMVQITPETMEGLRLALREQKDFRIQ 1357
> ath:AT5G44790 RAN1; RAN1 (RESPONSIVE-TO-ANTAGONIST 1); ATPase,
coupled to transmembrane movement of ions, phosphorylative
mechanism / copper ion transmembrane transporter; K01533 Cu2+-exporting
ATPase [EC:3.6.3.4]
Length=1001
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 49/92 (53%), Gaps = 11/92 (11%)
Query 22 KMVVAIKGILVQCDPPTME-IILQLNETKNFIIEKISEE-------HVLCSESVCSFLEE 73
K+V+ + GIL + D +E I+ +LN + F +++IS E V+ S S+ +EE
Sbjct 208 KLVLRVDGILNELDAQVLEGILTRLNGVRQFRLDRISGELEVVFDPEVVSSRSLVDGIEE 267
Query 74 EVTRRLELAERPLAEMQKEKKANEAGAAANTY 105
+ + +L R ++ ++ + + G A+N +
Sbjct 268 DGFGKFKL--RVMSPYER-LSSKDTGEASNMF 296
> ath:AT1G62886 DNA binding
Length=72
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 4/36 (11%)
Query 23 MVVAIKGILVQCDPPTMEIILQLNE----TKNFIIE 54
MV +IKG+ + CD P + I LN ++ FII+
Sbjct 1 MVNSIKGVFISCDVPMAQFIAHLNNSLPASQKFIIQ 36
> dre:100005449 zfyve9, si:ch211-57g18.2; zinc finger, FYVE domain
containing 9; K04679 MAD, mothers against decapentaplegic
interacting protein
Length=1209
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 12/63 (19%)
Query 11 FFIFK-----FGDFLAKMVVAIKGILVQCDPPTMEIILQ-LNETKNFII------EKISE 58
FF+F +LAK + G++VQ TME + Q L E K+F I ++ ++
Sbjct 986 FFVFSGALKASSGYLAKTSIVEDGVMVQITAETMEALRQALREMKDFSIACGKADQEENQ 1045
Query 59 EHV 61
EHV
Sbjct 1046 EHV 1048
> mmu:74068 Asz1, 4933400N19Rik, Gasz, ORF3; ankyrin repeat, SAM
and basic leucine zipper domain containing 1
Length=475
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 40/86 (46%), Gaps = 10/86 (11%)
Query 9 FFFFIFKFGDFLAKMVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESVC 68
F F+ K ++ A++ I+ + + +I+L+ +NF + EE V E
Sbjct 356 FLNFLLKLNKQCGHLITAVQNIITELPVNSHKIVLEWASPRNFT--SVCEELVSNVED-- 411
Query 69 SFLEEEVTRRLELAERPLAEMQKEKK 94
L EEV R EL ++ MQ E++
Sbjct 412 --LNEEVCRLKELIQK----MQNERE 431
> hsa:126402 CCDC105, FLJ40365; coiled-coil domain containing
105
Length=499
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 26/55 (47%), Gaps = 13/55 (23%)
Query 43 LQLNETKNFIIEK-------------ISEEHVLCSESVCSFLEEEVTRRLELAER 84
L LNE K ++E + E+ + S+ VC+ L ++ + LEL ER
Sbjct 295 LALNEAKRLLVESKDTLVEMAKNEVDVREQQLQISDRVCASLAQKASETLELKER 349
> dre:559694 ectodermal-neural cortex (with BTB-like domain)-like;
K10466 kelch-like protein 30
Length=650
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 29/63 (46%), Gaps = 3/63 (4%)
Query 8 HFFFFIFKFGDFLAKMVVAIKGILVQCDPPTMEIILQLNETKNFIIEKISEEHVLCSESV 67
H+F +F GDF+ + ++ L DP + +L T I K + E ++C+ S
Sbjct 56 HYFHSMFS-GDFIESIAARVE--LHDVDPDVLSSLLDFAYTGKLTINKNNVEGLICTSSQ 112
Query 68 CSF 70
F
Sbjct 113 LQF 115
> hsa:63967 CLSPN, MGC131612, MGC131613, MGC131615; claspin
Length=1275
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 17/59 (28%), Positives = 32/59 (54%), Gaps = 1/59 (1%)
Query 37 PTMEIILQLNETKNFIIE-KISEEHVLCSESVCSFLEEEVTRRLELAERPLAEMQKEKK 94
P +E+ LQ + +F + K S++H+ E + + RRLE ER + ++++ KK
Sbjct 130 PCLELSLQSGNSTDFTTDRKSSKKHIHDKEGTAGKAKVKSKRRLEKEERKMEKIRQLKK 188
> ath:AT1G50830 hypothetical protein
Length=768
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 29/46 (63%), Gaps = 5/46 (10%)
Query 51 FIIEKISEEHVLCSESVCSFL--EEEVTRRLELAERPLAEMQKEKK 94
F++ K SE+H C+E +CS + +EE+ RL+ ER LA + E K
Sbjct 695 FLVHKNSEKH-RCNEKMCSEVKKQEEIDERLK--ERKLAIKEMELK 737
> dre:100005596 hypothetical LOC100005596
Length=1698
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 47 ETKNFIIEKISEEHVLCSESVCSFLEEEVTRRLELAERP 85
E K ++K +EE C E +C FLEE+ TR +L+ P
Sbjct 990 EDKECALQKQAEELQKCQEKIC-FLEEKQTRTNDLSVPP 1027
> tpv:TP02_0649 hypothetical protein
Length=623
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Query 32 VQCDPPTMEIILQLNETKNFIIEKISEEHVLC-SESVCSFLEEEVTRRLEL 81
V C + I +QL +T + +I I ++ LC S V F+EE + +EL
Sbjct 7 VLCRSTSRTIRIQLIQTMSMLIHNIGKKKTLCESNDVLCFIEELMDLEIEL 57
> ath:AT4G18450 ethylene-responsive factor, putative
Length=303
Score = 27.7 bits (60), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query 52 IIEKISEEHVLCSESVCSFLEEEVTRRLELAERPLAEMQKEKKANE 97
+++ + E H C+ S CS E TR L +E +++K +++NE
Sbjct 217 VVDGMVENH--CALSYCSTKEHSETRGLRGSEETWFDLRKRRRSNE 260
Lambda K H
0.326 0.139 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017521068
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40