bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
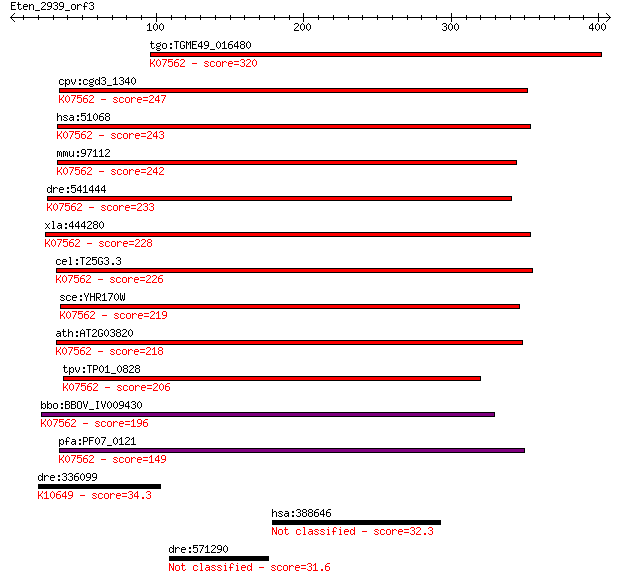
Query= Eten_2939_orf3
Length=407
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_016480 NMD3 protein ; K07562 nonsense-mediated mRNA... 320 8e-87
cpv:cgd3_1340 NMD3p like protein involved in nonsense mediated... 247 6e-65
hsa:51068 NMD3, FLJ21053; NMD3 homolog (S. cerevisiae); K07562... 243 1e-63
mmu:97112 Nmd3, C87860; NMD3 homolog (S. cerevisiae); K07562 n... 242 2e-63
dre:541444 nmd3, nmd3l, zgc:113103; NMD3 homolog (S. cerevisia... 233 8e-61
xla:444280 nmd3, MGC80914, nmd3l; NMD3 homolog; K07562 nonsens... 228 3e-59
cel:T25G3.3 hypothetical protein; K07562 nonsense-mediated mRN... 226 9e-59
sce:YHR170W NMD3, SRC5; Nmd3p; K07562 nonsense-mediated mRNA d... 219 2e-56
ath:AT2G03820 nonsense-mediated mRNA decay NMD3 family protein... 218 4e-56
tpv:TP01_0828 hypothetical protein; K07562 nonsense-mediated m... 206 9e-53
bbo:BBOV_IV009430 23.m05734; nonsense-mediated mRNA decay prot... 196 2e-49
pfa:PF07_0121 60S ribosomal subunit export protein, putative; ... 149 2e-35
dre:336099 trim9, fj48c11, wu:fj48c11, zgc:77357; tripartite m... 34.3 0.88
hsa:388646 GBP7, FLJ38822, GBP4L; guanylate binding protein 7 32.3
dre:571290 si:dkey-78d16.2 31.6 5.8
> tgo:TGME49_016480 NMD3 protein ; K07562 nonsense-mediated mRNA
decay protein 3
Length=721
Score = 320 bits (819), Expect = 8e-87, Method: Compositional matrix adjust.
Identities = 152/307 (49%), Positives = 218/307 (71%), Gaps = 9/307 (2%)
Query 96 RELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSFNVSG 155
REL+A+CLK++RGLK + +I DC+WL++EPHS ++ ++LT ++DVL+G+A ++QS + G
Sbjct 12 RELMAVCLKKVRGLKKVEKIVDCQWLWTEPHSRRVKMKLTVQSDVLDGRAGLQQSLIIEG 71
Query 156 LVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRVVQRR 215
++ N QCDECK+++TPHAGW A VQVRQ +HKRTFLFLEQL+L+ + L +LL VV++
Sbjct 72 VIQNTQCDECKKSYTPHAGWTAMVQVRQHAEHKRTFLFLEQLVLKHR-LAARLLNVVEKS 130
Query 216 DGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPAICRD 275
DGLDF+FP++QAA F FC SR P SR +KQLISHD +SN Y++K+T+ V L +C+D
Sbjct 131 DGLDFHFPSRQAALHFADFCQSRFPSVSRQAKQLISHDASSNIYYHKYTISVTLAPVCKD 190
Query 276 DLVYIPAKKAQDSFGGFPSLSLCTRVSST-LHLTDPFSGRHLDMAADVYFRDTFKPICTR 334
DLVYI K+AQ GGF +LSLCTRV+ + LTDPFSG+ L+++ + Y++ F+P+CTR
Sbjct 191 DLVYIHRKQAQAQLGGFTNLSLCTRVAGAGIQLTDPFSGKVLNVSPEKYWKSPFQPLCTR 250
Query 335 KHLSHFLIVAVTPLQQTQQKQPQQKQQHGRRHQRQQNKQQQQQEAAAGDDAMTVDTSAEA 394
KHLS FL++ V P+ + Q + RR ++ K + + GD+ D A
Sbjct 251 KHLSSFLVLDVNPVDRPGYSQTESGS---RRDVPRKGKGRATRSEPEGDE----DAEMFA 303
Query 395 GGAGTGR 401
G TGR
Sbjct 304 GLQTTGR 310
> cpv:cgd3_1340 NMD3p like protein involved in nonsense mediated
decay ; K07562 nonsense-mediated mRNA decay protein 3
Length=588
Score = 247 bits (630), Expect = 6e-65, Method: Compositional matrix adjust.
Identities = 129/324 (39%), Positives = 203/324 (62%), Gaps = 12/324 (3%)
Query 34 HVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQGKWLSCEL 93
++ CC+CG T +P C++C++ E+D++ + R ++ CRQC RF+ + W+SC+L
Sbjct 44 YMNCCVCGVATEINPSRMCINCIRSEVDITEGISRQAIISFCRQCERFQ--KPPWVSCQL 101
Query 94 ESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSFNV 153
ESRELLALCLK+I+GL ++ R+ D ++++EPHS +L ++ T + +V+ G A ++QS V
Sbjct 102 ESRELLALCLKKIKGLNTV-RLVDASFIWTEPHSRRLKVKCTIQKEVMNG-AIIQQSIVV 159
Query 154 SGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRVVQ 213
+ QCD+C+++FTPH W VQVRQR +HKRTFL+LEQL+L+ +K+ +V+
Sbjct 160 EFFMQAQQCDDCRRSFTPHT-WNTVVQVRQRVEHKRTFLYLEQLILKH-GAHEKVSNIVE 217
Query 214 RRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPAIC 273
+ DGLDF F K A V F + ++S+QLIS D +SNT+ K T VEL IC
Sbjct 218 KPDGLDFQFQQKSHAQKLVDFITHYFASKVKNSRQLISQDLSSNTHNNKFTFSVELCPIC 277
Query 274 RDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVYFR------DT 327
+DD+V+IP GG SL LC ++++ + L DPF+GR D+ + Y++ +
Sbjct 278 KDDVVFIPKNLCISLLGGISSLLLCKKITNRISLIDPFTGREADLTKEKYWQYSGSNGSS 337
Query 328 FKPICTRKHLSHFLIVAVTPLQQT 351
F P+ +R L F ++ V + T
Sbjct 338 FIPLLSRSSLVDFYVLNVERINTT 361
> hsa:51068 NMD3, FLJ21053; NMD3 homolog (S. cerevisiae); K07562
nonsense-mediated mRNA decay protein 3
Length=503
Score = 243 bits (620), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 123/321 (38%), Positives = 195/321 (60%), Gaps = 4/321 (1%)
Query 33 GHVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQGKWLSCE 92
GH+LCC CG P+P N C++CL+ ++D+S + + + C+QC R+ G W+ C
Sbjct 13 GHILCCECGVPISPNPANICVACLRSKVDISQGIPKQVSISFCKQCQRYFQPPGTWIQCA 72
Query 93 LESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSFN 152
LESRELLALCLK+I+ S R+ D ++++EPHS +L ++LT + +V+ G A ++Q F
Sbjct 73 LESRELLALCLKKIKAPLSKVRLVDAGFVWTEPHSKRLKVKLTIQKEVMNG-AILQQVFV 131
Query 153 VSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRVV 212
V +V + C +C + W A +QVRQ+ HK+TF +LEQL+L+ + Q LR+
Sbjct 132 VDYVVQSQMCGDCHRV-EAKDFWKAVIQVRQKTLHKKTFYYLEQLILKYG-MHQNTLRIK 189
Query 213 QRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPAI 272
+ DGLDFY+ +KQ A V F +PC ++S++LIS D +SNTY YK T VE+ I
Sbjct 190 EIHDGLDFYYSSKQHAQKMVEFLQCTVPCRYKASQRLISQDIHSNTYNYKSTFSVEIVPI 249
Query 273 CRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVYFRDTFKPIC 332
C+D++V + K AQ S G + +C RV+S +HL DP + + D+ ++ F +C
Sbjct 250 CKDNVVCLSPKLAQ-SLGNMNQICVCIRVTSAIHLIDPNTLQVADIDGSTFWSHPFNSLC 308
Query 333 TRKHLSHFLIVAVTPLQQTQQ 353
K L F+++ + +Q ++
Sbjct 309 HPKQLEEFIVMECSIVQDIKR 329
> mmu:97112 Nmd3, C87860; NMD3 homolog (S. cerevisiae); K07562
nonsense-mediated mRNA decay protein 3
Length=503
Score = 242 bits (617), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 122/311 (39%), Positives = 190/311 (61%), Gaps = 4/311 (1%)
Query 33 GHVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQGKWLSCE 92
GH+LCC CG P+P N C++CL+ ++D+S V + + C+QC R+ W+ C
Sbjct 13 GHILCCECGVPISPNPANICVACLRSKVDISQGVPKQVSISFCKQCQRYFQPPASWVQCA 72
Query 93 LESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSFN 152
LESRELLALCLK+I+ S R+ D ++++EPHS +L ++LT + +V+ G A ++Q F
Sbjct 73 LESRELLALCLKKIKAPLSKVRLVDAGFVWTEPHSKRLKVKLTIQKEVMNG-AILQQVFV 131
Query 153 VSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRVV 212
V +V + C +C + W A +QVRQ+ HK+TF +LEQL+L+ + Q LR+
Sbjct 132 VDYVVQSQMCGDCHRV-EAKDFWKAVIQVRQKTLHKKTFYYLEQLILKYG-MHQNTLRIK 189
Query 213 QRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPAI 272
+ DGLDFY+ +KQ A V F +PC ++S++LIS D +SNTY YK T VE+ I
Sbjct 190 EIHDGLDFYYSSKQHAQKMVEFLQGIVPCRYKASQRLISQDIHSNTYNYKSTFSVEIVPI 249
Query 273 CRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVYFRDTFKPIC 332
C+D++V + K AQ S G + +C RV+S +HL DP + + D+ + ++ F +C
Sbjct 250 CKDNVVCLSPKLAQ-SLGNMSQICICIRVTSAIHLIDPNTLQVADIDGNTFWSHPFNSLC 308
Query 333 TRKHLSHFLIV 343
K L F+++
Sbjct 309 HPKQLEEFIVI 319
> dre:541444 nmd3, nmd3l, zgc:113103; NMD3 homolog (S. cerevisiae);
K07562 nonsense-mediated mRNA decay protein 3
Length=505
Score = 233 bits (595), Expect = 8e-61, Method: Compositional matrix adjust.
Identities = 124/315 (39%), Positives = 196/315 (62%), Gaps = 4/315 (1%)
Query 26 AGPSVGVGHVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQ 85
A PS G++LCC CG PP+P N C+SCL+ ++D+S + + + CRQC R+
Sbjct 6 APPSSSHGNILCCTCGVPIPPNPANMCVSCLRTQVDISEGIPKQVTIHFCRQCERYLQPP 65
Query 86 GKWLSCELESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKA 145
G W+ C LESRELLALCLK+++ S R+ D +L++EPHS ++ ++LT + +V+ G A
Sbjct 66 GSWVQCALESRELLALCLKKLKASMSKVRLIDAGFLWTEPHSKRIKIKLTIQKEVMNG-A 124
Query 146 EVEQSFNVSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLL 205
++Q F V +V C++C + W A VQVRQ+ HK+TF +LEQL+L+ + L
Sbjct 125 ILQQVFTVEFVVQGQMCEDCHRV-EAKDFWKAVVQVRQKTVHKKTFYYLEQLILKHK-LH 182
Query 206 QKLLRVVQRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTV 265
Q +R+ + +G+DFY+ KQ A V F +PC S++S++LISHD +SNTY YK T
Sbjct 183 QNTVRIKEIHEGIDFYYATKQHAQKMVDFLQCTVPCRSKASQRLISHDVHSNTYNYKSTF 242
Query 266 YVELPAICRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVYFR 325
VE+ +C+D++V + + AQ S G + +C RV+S++HL DP + + ++ ++R
Sbjct 243 SVEIVPVCKDNVVCLSPRLAQ-SLGNMGQVCVCVRVTSSIHLIDPNTLQIAEVDGSTFWR 301
Query 326 DTFKPICTRKHLSHF 340
F +C+ + L F
Sbjct 302 HPFNSLCSPRQLEEF 316
> xla:444280 nmd3, MGC80914, nmd3l; NMD3 homolog; K07562 nonsense-mediated
mRNA decay protein 3
Length=504
Score = 228 bits (582), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 126/330 (38%), Positives = 199/330 (60%), Gaps = 5/330 (1%)
Query 25 MAGPSVGV-GHVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKD 83
M GP+ G++LCC CG PP+P N C++CL+ ++D+S + + V C+QC R+
Sbjct 4 MKGPATSSQGNILCCQCGIPIPPNPANMCVACLRTQVDISEGIPKQVSVHFCKQCERYLQ 63
Query 84 SQGKWLSCELESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEG 143
G W+ C LESRELL LCLK+++ S R+ D ++++EPHS +L ++LT + +V+ G
Sbjct 64 PPGTWIQCALESRELLTLCLKKLKANLSKVRLIDAGFIWTEPHSKRLKVKLTIQKEVMNG 123
Query 144 KAEVEQSFNVSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQH 203
A ++Q F V +V CD+C + W A VQVRQ+ HK+TF +LEQ +L+ +
Sbjct 124 -AILQQVFVVDYVVQAQMCDDCHRV-EAKDFWKAVVQVRQKVLHKKTFYYLEQTILKHR- 180
Query 204 LLQKLLRVVQRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKH 263
L Q LRV + DGLDFY+ +KQ V F +PC ++S++LISHD ++N Y YK
Sbjct 181 LHQNTLRVKEIHDGLDFYYASKQHGQKMVEFLQCTVPCRFKASQRLISHDIHTNAYNYKS 240
Query 264 TVYVELPAICRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVY 323
T VE+ +C+D+LV + K AQ S G + +C RV+ST+HL DP + + ++ + Y
Sbjct 241 TFSVEIVPVCKDNLVCLSPKLAQ-SLGNMNQICICARVTSTIHLIDPSTLQIAEVDGNTY 299
Query 324 FRDTFKPICTRKHLSHFLIVAVTPLQQTQQ 353
+R F + K + F+++ ++ +Q
Sbjct 300 WRYPFNSLFHPKQMEEFIVMDTEIIRDRKQ 329
> cel:T25G3.3 hypothetical protein; K07562 nonsense-mediated mRNA
decay protein 3
Length=513
Score = 226 bits (577), Expect = 9e-59, Method: Compositional matrix adjust.
Identities = 117/324 (36%), Positives = 192/324 (59%), Gaps = 6/324 (1%)
Query 32 VGHVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQGKWLSC 91
VG + CC CG P+P N C CL+ +D++ + R + C+ C R+ W+
Sbjct 10 VGLIACCECGVAIEPNPANMCSGCLRSRVDITEGITRQCTIYMCKFCDRYFVPPSAWMRA 69
Query 92 ELESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSF 151
ELES+ELL++CLK+++ + + R++D ++++E HS ++ +++T + +V ++Q+
Sbjct 70 ELESKELLSICLKKLKPMLTKVRLTDACFVWTEAHSKRIKVKITIQKEVFTNTI-LQQAV 128
Query 152 NVSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRV 211
V VH+ CD+C++A W A VQVRQR + K+T +LEQLLL +H K
Sbjct 129 VVEFTVHSQLCDDCRRA-EAKDFWRACVQVRQRAEFKKTLFYLEQLLL--KHSAHKECTG 185
Query 212 VQR-RDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELP 270
V+ G+DFYF +Q A FV F + LPC +++L+SHD +NTY YKHT VE+
Sbjct 186 VKPVPTGIDFYFAKQQEARKFVDFLMTVLPCKYHYAQELVSHDTKNNTYDYKHTFCVEIV 245
Query 271 AICRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVYFRDTFKP 330
ICRD++V +P K+AQ +G + LC RVS+ + L DP + + +D+ A ++R+ F
Sbjct 246 PICRDNIVCLPKKQAQ-QYGNMSQIVLCLRVSNVITLIDPNNLQLVDVQATNFWREPFDS 304
Query 331 ICTRKHLSHFLIVAVTPLQQTQQK 354
+C K L+ F ++ V P+ ++K
Sbjct 305 LCGPKQLTEFYVLDVEPVNNFERK 328
> sce:YHR170W NMD3, SRC5; Nmd3p; K07562 nonsense-mediated mRNA
decay protein 3
Length=518
Score = 219 bits (557), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 110/314 (35%), Positives = 185/314 (58%), Gaps = 10/314 (3%)
Query 35 VLCCICGAVTPPHPRN---KCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQGKWLSC 91
+LCC CG TP C C++ +D++ + R + CR C RF G+W+
Sbjct 17 LLCCNCG--TPIDGSTGLVMCYDCIKLTVDITQGIPREANISFCRNCERFLQPPGQWIRA 74
Query 92 ELESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSF 151
ELESRELLA+CL+R++GL + R+ D ++++EPHS ++ ++LT + + + ++Q+F
Sbjct 75 ELESRELLAICLRRLKGLTKV-RLVDASFIWTEPHSRRIRIKLTVQGEAMTNTI-IQQTF 132
Query 152 NVSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRV 211
V +V MQC +C +++T + W A VQ+RQ+ HKRTFLFLEQL+L+ + + +
Sbjct 133 EVEYIVIAMQCPDCARSYTTNT-WRATVQIRQKVPHKRTFLFLEQLILKHNAHVDTI-SI 190
Query 212 VQRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPA 271
+ +DGLDF++ K A + F ++ +P + S++LIS D ++ YK + VE+
Sbjct 191 SEAKDGLDFFYAQKNHAVKMIDFLNAVVPIKHKKSEELISQDTHTGASTYKFSYSVEIVP 250
Query 272 ICRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVYFRDTFKPI 331
IC+DDLV +P K A+ S G LC+++S+T+ DP + + D++ VY+R F +
Sbjct 251 ICKDDLVVLPKKLAK-SMGNISQFVLCSKISNTVQFMDPTTLQTADLSPSVYWRAPFNAL 309
Query 332 CTRKHLSHFLIVAV 345
L F+++ V
Sbjct 310 ADVTQLVEFIVLDV 323
> ath:AT2G03820 nonsense-mediated mRNA decay NMD3 family protein;
K07562 nonsense-mediated mRNA decay protein 3
Length=516
Score = 218 bits (555), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 111/316 (35%), Positives = 182/316 (57%), Gaps = 4/316 (1%)
Query 32 VGHVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQGKWLSC 91
+G VLCC CG P+ N C++CL+ E+D++ + ++ + C +C + W+ C
Sbjct 16 IGSVLCCKCGVPMAPNAANMCVNCLRSEVDITEGLQKSIQIFYCPECTCYLQPPKTWIKC 75
Query 92 ELESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSF 151
+ ES+ELL C+KR++ L + ++ + ++++EPHS ++ ++LT +A+VL G A +EQS+
Sbjct 76 QWESKELLTFCIKRLKNLNKV-KLKNAEFVWTEPHSKRIKVKLTVQAEVLNG-AVLEQSY 133
Query 152 NVSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRV 211
V V + C+ C + W A +Q+RQ H+RTF +LEQL+L+ + +R+
Sbjct 134 PVEYTVRDNLCESCSRFQANPDQWVASIQLRQHVSHRRTFFYLEQLILRHDAA-SRAIRI 192
Query 212 VQRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPA 271
Q G+DF+F NK A SFV F +P R +QL+SHD S+ Y YK+T V++
Sbjct 193 QQVDQGIDFFFGNKSHANSFVEFLRKVVPIEYRQDQQLVSHDVKSSLYNYKYTYSVKICP 252
Query 272 ICRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMAADVYFRDTFKPI 331
+CR+DLV +P+K A G L +CT+VS + L DP + R + A Y+R F+
Sbjct 253 VCREDLVCLPSKVAS-GLGNLGPLVVCTKVSDNITLLDPRTLRCAFLDARQYWRSGFRSA 311
Query 332 CTRKHLSHFLIVAVTP 347
T + L + + V P
Sbjct 312 LTSRQLVKYFVFDVEP 327
> tpv:TP01_0828 hypothetical protein; K07562 nonsense-mediated
mRNA decay protein 3
Length=491
Score = 206 bits (525), Expect = 9e-53, Method: Compositional matrix adjust.
Identities = 107/284 (37%), Positives = 171/284 (60%), Gaps = 8/284 (2%)
Query 37 CCICGA-VTPPHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGRFKDSQGKWLSCELES 95
C +CG V+ P C +CL +DLS V + +C C +F +WLSCELES
Sbjct 21 CFLCGEFVSAPTSSRMCSTCLTKSVDLSENVSTHCTILQCVTCTKF--YHDRWLSCELES 78
Query 96 RELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSFNVSG 155
+ELL++CLK+I+G+ L +I D + ++EPHS KL + +T + +V E VEQS ++
Sbjct 79 KELLSVCLKKIKGI-DLMKIVDASFKWTEPHSKKLKINVTVQKEV-ENNCVVEQSLSIDF 136
Query 156 LVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRVVQRR 215
+ + QC CKQ +TPH W A VQ+RQ+ K+ L LEQ++L+ + + +L ++ RR
Sbjct 137 TIVSTQCASCKQLYTPHT-WKALVQIRQKTKDKKHILLLEQIILKH-NAHENVLNILSRR 194
Query 216 DGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPAICRD 275
+G D +F ++ A F F S ++ ++SK+L++ D N+N Y YK+T++V L IC D
Sbjct 195 NGFDLHFSSRNDAQKFADFVSDKIVSQIKNSKKLVTQDVNNNKYNYKYTLHVSLVPICTD 254
Query 276 DLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPFSGRHLDMA 319
DLV++ + K +GG L ++ST+ L DPF+ R L+++
Sbjct 255 DLVFL-SPKVSSIYGGISPFLLVVNLTSTISLMDPFTLRRLEVS 297
> bbo:BBOV_IV009430 23.m05734; nonsense-mediated mRNA decay protein;
K07562 nonsense-mediated mRNA decay protein 3
Length=549
Score = 196 bits (497), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 105/319 (32%), Positives = 180/319 (56%), Gaps = 19/319 (5%)
Query 22 AVAMAGPSVGVGHVLCCICGAVTP-PHPRNKCLSCLQDEIDLSSRVCRTFVVDRCRQCGR 80
A A + CC+CG V P P C C+ ++L+ V + +C C
Sbjct 6 AATFASTEPSYAPMFCCLCGDVVPKPTASRMCHLCITKNVNLAQEVTTHTTILQCGTCKM 65
Query 81 FKDSQGKWLSCELESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADV 140
F +WL C+LES+ELL++CLK+ +GL+++ +I + + ++EPHS +L +++ + +V
Sbjct 66 F--FHDRWLKCDLESKELLSICLKKAKGLENM-KIINAAFSWTEPHSKQLKVKVCVQKEV 122
Query 141 LEGKAEVEQSFNVSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQ 200
+ VEQSF ++ + + QCD CK+ +TPH W A VQ+RQ+ KR L LEQ++++
Sbjct 123 -DSNFVVEQSFVITFSIKSSQCDHCKRMYTPHT-WKAMVQIRQKTKDKRHILMLEQIIIK 180
Query 201 QQHLLQKLLRVVQRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYF 260
+ + + ++ R +G D +F +K A F F S ++ R+SK+LI+ D +N +
Sbjct 181 H-NAHENVTNILSRPNGFDLHFQSKTCAQKFADFVSDKIVSQVRNSKKLITQDSVNNKWD 239
Query 261 YKHTVYVELPAICRDDL-----------VYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTD 309
YK+T+ V+L IC DD+ V++P K A + GG LC +++T+ L D
Sbjct 240 YKYTLQVQLIPICADDMVRANNDVNISQVFLPPKVASIN-GGISPFLLCVNLTTTISLID 298
Query 310 PFSGRHLDMAADVYFRDTF 328
PF+ R LD++ D ++++ F
Sbjct 299 PFTLRTLDISNDKFWKNPF 317
> pfa:PF07_0121 60S ribosomal subunit export protein, putative;
K07562 nonsense-mediated mRNA decay protein 3
Length=888
Score = 149 bits (377), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 99/319 (31%), Positives = 179/319 (56%), Gaps = 10/319 (3%)
Query 34 HVLCCICGAVTPPHPRNKCLSCLQDEIDLSSRVCR--TFVVDRCRQCGRFKDSQGKWLSC 91
++ C +CG + C +C+ I+ +S T+++ CR+C R+ +W+ C
Sbjct 320 YISCILCGDSIKANTSKMCHNCILQNIESNSININKDTYLIYYCRECKRY--LHNRWVYC 377
Query 92 ELESRELLALCLKRIRGLKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSF 151
ELES+ELLALCLK++ LK L + ++LY+EPHS +L + ++ + +++ E
Sbjct 378 ELESKELLALCLKKVNKLKKLKILD-AKFLYTEPHSKRLKVHVSVQEELINNFIS-EMEI 435
Query 152 NVSGLVHNMQCDECKQAFTPHAGWGARVQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRV 211
+ ++ QCD+CK+ +TP+ + V VRQ+ +HK+T L + LL + ++ + ++ +
Sbjct 436 IIHYVIKYTQCDDCKKKYTPYT-YNTCVSVRQKVEHKKTLL-FLENLLLKCNMNENIINI 493
Query 212 VQRRDGLDFYFPNKQAAASFVSFCSSRLPCSSRSSKQLISHDCNSNTYFYKHTVYVELPA 271
V DGLDF+F ++ A F F S+ ++SK LI+HD N+NTY Y +T +++
Sbjct 494 VSNPDGLDFHFLSRTDALKFCDFILSKTMSKCKNSKHLINHDANNNTYNYLYTFSIDICP 553
Query 272 ICRDDLVYIPAKKAQDSFGGFPSLSLCTRVSSTLHLTDPF-SGRHLDMAADVYFRDTFKP 330
IC+ DL++ P K + +G LC VS + L +PF S H+ ++ + Y ++ F P
Sbjct 554 ICKYDLIFFP-KGLANKYGIKNVFYLCIHVSIFIILINPFSSANHVYVSPERYNKNPFLP 612
Query 331 ICTRKHLSHFLIVAVTPLQ 349
+ ++ FLI+ + +
Sbjct 613 LLSKADAKVFLILNIEYIN 631
> dre:336099 trim9, fj48c11, wu:fj48c11, zgc:77357; tripartite
motif-containing 9; K10649 tripartite motif-containing protein
9/67
Length=699
Score = 34.3 bits (77), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 39/90 (43%), Gaps = 9/90 (10%)
Query 20 AFAVAMAGPSVGVGHVLCCICGAVTPPHPRNKCLSCLQ--DEIDLSSRVCRTFVVDRCRQ 77
F +M P H+L V PP PRN CL+C Q + L R R F +R +
Sbjct 101 VFPPSMPPPQQAQHHLLPHT--GVLPPVPRNSCLTCPQCHRSLTLDDRGLRGFPKNRVLE 158
Query 78 --CGRFKDSQGKWLSCEL---ESRELLALC 102
R++ S+ L C+L RE +C
Sbjct 159 GVVDRYQQSKAAALKCQLCEKSPREATVMC 188
> hsa:388646 GBP7, FLJ38822, GBP4L; guanylate binding protein
7
Length=638
Score = 32.3 bits (72), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 26/118 (22%), Positives = 48/118 (40%), Gaps = 11/118 (9%)
Query 179 VQVRQRCDHKRTFLFLEQLLLQQQHLLQKLLRVVQRRDGLDFYFPNKQAAASFVSFCSSR 238
+ V C+ + +F+E + QK L + DF N++A+A +C +
Sbjct 353 LDVHAVCEREAIAVFMEYSFKDKSQEFQKKLVDTMEKKKEDFVLQNEEASA---KYCQAE 409
Query 239 LPCSSRSSKQLISHDCNSNTYFYK--HTVYVELPAICRDDLVYIPAK--KAQDSFGGF 292
L + +L++ + T+F H +Y+E D +P K KA + F
Sbjct 410 L----KRLSELLTESISRGTFFVPGGHNIYLEAKKKIEQDYTLVPRKGVKADEVLQSF 463
> dre:571290 si:dkey-78d16.2
Length=809
Score = 31.6 bits (70), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 30/67 (44%), Gaps = 0/67 (0%)
Query 109 LKSLSRISDCRWLYSEPHSNKLLLRLTARADVLEGKAEVEQSFNVSGLVHNMQCDECKQA 168
LK + I + ++ +P K LL ++ VL G +E S VS H C EC A
Sbjct 417 LKKGAHIIEEIQVFHQPQMVKSLLLSVSKGAVLIGSSEGLLSVPVSNCSHYRSCAECVLA 476
Query 169 FTPHAGW 175
P GW
Sbjct 477 RDPFCGW 483
Lambda K H
0.324 0.134 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 18441069616
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40