bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2936_orf2
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
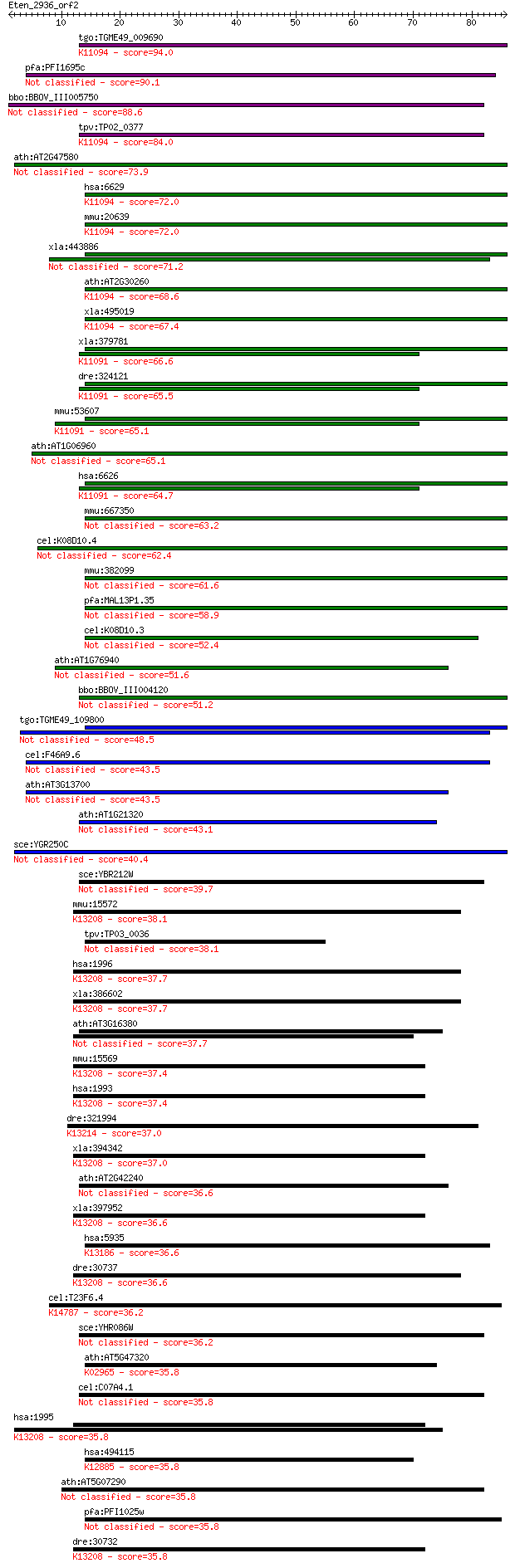
tgo:TGME49_009690 U2 small nuclear ribonucleoprotein, putative... 94.0 1e-19
pfa:PFI1695c small nuclear ribonucleoprotein (snRNP), putative 90.1 1e-18
bbo:BBOV_III005750 17.m07512; U2 small nuclear ribonucleoprote... 88.6 4e-18
tpv:TP02_0377 U2 small nuclear ribonucleoprotein B; K11094 U2 ... 84.0 1e-16
ath:AT2G47580 U1A; U1A (SPLICEOSOMAL PROTEIN U1A); RNA binding... 73.9 1e-13
hsa:6629 SNRPB2, MGC24807, MGC45309, Msl1; small nuclear ribon... 72.0 4e-13
mmu:20639 Snrpb2, 2810052G09Rik; U2 small nuclear ribonucleopr... 72.0 5e-13
xla:443886 MGC80122 protein 71.2 7e-13
ath:AT2G30260 U2B''; U2B'' (U2 small nuclear ribonucleoprotein... 68.6 5e-12
xla:495019 snrpb2; small nuclear ribonucleoprotein polypeptide... 67.4 1e-11
xla:379781 snrpa, MGC52976, U1A, snf; small nuclear ribonucleo... 66.6 2e-11
dre:324121 snrpa, fc19d01, wu:fc19d01, zgc:77810; small nuclea... 65.5 4e-11
mmu:53607 Snrpa, C430021M15Rik, Rnu1a-1, Rnu1a1, U1A; small nu... 65.1 6e-11
ath:AT1G06960 small nuclear ribonucleoprotein U2B, putative / ... 65.1 6e-11
hsa:6626 SNRPA, Mud1, U1-A, U1A; small nuclear ribonucleoprote... 64.7 7e-11
mmu:667350 Gm8587, EG667350; predicted pseudogene 8587 63.2
cel:K08D10.4 rnp-2; RNP (RRM RNA binding domain) containing fa... 62.4 3e-10
mmu:382099 Gm5161, EG382099; predicted pseudogene 5161 61.6
pfa:MAL13P1.35 U1 small nuclear ribonucleoprotein A, putative 58.9 4e-09
cel:K08D10.3 rnp-3; RNP (RRM RNA binding domain) containing fa... 52.4 3e-07
ath:AT1G76940 RNA recognition motif (RRM)-containing protein 51.6 6e-07
bbo:BBOV_III004120 17.m07378; U2 small nuclear ribonucleoprote... 51.2 7e-07
tgo:TGME49_109800 small nuclear ribonucleoprotein U1A, putative 48.5 5e-06
cel:F46A9.6 mec-8; MEChanosensory abnormality family member (m... 43.5 2e-04
ath:AT3G13700 RNA-binding protein, putative 43.5 2e-04
ath:AT1G21320 nucleic acid binding / nucleotide binding 43.1 2e-04
sce:YGR250C Putative RNA binding protein; localizes to stress ... 40.4 0.002
sce:YBR212W NGR1, RBP1; Ngr1p 39.7 0.002
mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abn... 38.1 0.007
tpv:TP03_0036 hypothetical protein 38.1 0.007
hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal v... 37.7 0.009
xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic le... 37.7 0.009
ath:AT3G16380 PAB6; PAB6 (POLY(A) BINDING PROTEIN 6); RNA bind... 37.7 0.010
mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnorma... 37.4 0.013
hsa:1993 ELAVL2, HEL-N1, HELN1, HUB; ELAV (embryonic lethal, a... 37.4 0.013
dre:321994 nono, wu:fb40d10, wu:fc11d05; non-POU domain contai... 37.0 0.014
xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,... 37.0 0.017
ath:AT2G42240 nucleic acid binding / nucleotide binding 36.6 0.017
xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1; ... 36.6 0.017
hsa:5935 RBM3, IS1-RNPL, RNPL; RNA binding motif (RNP1, RRM) p... 36.6 0.021
dre:30737 elavl4, elrd, wu:fc24h01, zHuD; ELAV (embryonic leth... 36.6 0.021
cel:T23F6.4 rbd-1; RBD (RNA binding domain) protein family mem... 36.2 0.025
sce:YHR086W NAM8, MRE2, MUD15; Nam8p 36.2 0.025
ath:AT5G47320 RPS19; RPS19 (RIBOSOMAL PROTEIN S19); RNA bindin... 35.8 0.030
cel:C07A4.1 hypothetical protein 35.8 0.031
hsa:1995 ELAVL3, DKFZp547J036, HUC, HUCL, MGC20653, PLE21; ELA... 35.8 0.032
hsa:494115 RBMXL1, DKFZp547N1117, DKFZp667D0223, FLJ32614, RBM... 35.8 0.032
ath:AT5G07290 AML4; AML4 (ARABIDOPSIS MEI2-LIKE 4); RNA bindin... 35.8 0.032
pfa:PFI1025w RNA binding protein, putative 35.8 0.032
dre:30732 elavl3, HuC, elrc, id:ibd1248, wu:fb77b03, zHuC; ELA... 35.8 0.033
> tgo:TGME49_009690 U2 small nuclear ribonucleoprotein, putative
; K11094 U2 small nuclear ribonucleoprotein B''
Length=233
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 42/73 (57%), Positives = 55/73 (75%), Gaps = 0/73 (0%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKI 72
SLF+ENL +K +L+ILF QY+G E RL+EGRGVAFVDF+ A A VA+ G+QGFK+
Sbjct 161 SLFIENLPPKATKTSLDILFGQYRGHTESRLIEGRGVAFVDFSTQAQAAVAMQGMQGFKV 220
Query 73 SSSHQLRISHVER 85
SH ++IS VE+
Sbjct 221 DPSHPIKISMVEK 233
> pfa:PFI1695c small nuclear ribonucleoprotein (snRNP), putative
Length=193
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/84 (47%), Positives = 58/84 (69%), Gaps = 4/84 (4%)
Query 4 SKGALEARA----SLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAA 59
SK +E ++ +L V+NL + +K ALEILF+QY GF EVR + G+ +AFVDF A
Sbjct 109 SKHNIEHKSNLIYTLLVQNLPDEINKNALEILFNQYPGFYEVRYIPGKNIAFVDFTAQEH 168
Query 60 AEVALSGLQGFKISSSHQLRISHV 83
AE++++GLQ FKI+ H ++IS V
Sbjct 169 AEISMTGLQNFKITPHHPMKISWV 192
> bbo:BBOV_III005750 17.m07512; U2 small nuclear ribonucleoprotein
B'
Length=213
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 44/81 (54%), Positives = 55/81 (67%), Gaps = 2/81 (2%)
Query 1 DGTSKGALEARASLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAA 60
D S+G R +LFVENL +K ++E+LF QY G+++ R VEG+ VAFVDFA
Sbjct 128 DANSEGG--ERHTLFVENLHPDMNKMSVELLFQQYPGYKDTRFVEGKCVAFVDFATAYQG 185
Query 61 EVALSGLQGFKISSSHQLRIS 81
EVAL GLQGF +S SH LRIS
Sbjct 186 EVALQGLQGFSVSHSHALRIS 206
> tpv:TP02_0377 U2 small nuclear ribonucleoprotein B; K11094 U2
small nuclear ribonucleoprotein B''
Length=206
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 37/69 (53%), Positives = 51/69 (73%), Gaps = 0/69 (0%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKI 72
+LFV+N+ SK +LE+LF QY GFR R +EGR VAFVD++ + AE+AL GL GF++
Sbjct 135 ALFVQNIPHDMSKESLELLFKQYPGFRGCRFIEGRFVAFVDYSMASQAEIALEGLNGFRV 194
Query 73 SSSHQLRIS 81
S +H L+IS
Sbjct 195 SHTHALQIS 203
> ath:AT2G47580 U1A; U1A (SPLICEOSOMAL PROTEIN U1A); RNA binding
/ nucleic acid binding / nucleotide binding
Length=250
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/89 (42%), Positives = 57/89 (64%), Gaps = 6/89 (6%)
Query 2 GTSKGALEARAS----LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAA 56
G EA A LFV+NL T+ L++LF QY+GF+EVR++E + G+AFV+FA
Sbjct 163 GMKPNMPEAPAPPNNILFVQNLPHETTPMVLQMLFCQYQGFKEVRMIEAKPGIAFVEFAD 222
Query 57 PAAAEVALSGLQGFKISSSHQLRISHVER 85
+ VA+ GLQGFKI + Q+ I++ ++
Sbjct 223 EMQSTVAMQGLQGFKIQQN-QMLITYAKK 250
> hsa:6629 SNRPB2, MGC24807, MGC45309, Msl1; small nuclear ribonucleoprotein
polypeptide B; K11094 U2 small nuclear ribonucleoprotein
B''
Length=225
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 50/73 (68%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 153 LFLNNLPEETNEMMLSMLFNQFPGFKEVRLVPGRHDIAFVEFENDGQAGAARDALQGFKI 212
Query 73 SSSHQLRISHVER 85
+ SH ++I++ ++
Sbjct 213 TPSHAMKITYAKK 225
> mmu:20639 Snrpb2, 2810052G09Rik; U2 small nuclear ribonucleoprotein
B; K11094 U2 small nuclear ribonucleoprotein B''
Length=225
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 50/73 (68%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 153 LFLNNLPEETNEMMLSMLFNQFPGFKEVRLVPGRHDIAFVEFENDGQAGAARDALQGFKI 212
Query 73 SSSHQLRISHVER 85
+ SH ++I++ ++
Sbjct 213 TPSHAMKITYAKK 225
> xla:443886 MGC80122 protein
Length=223
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 34/73 (46%), Positives = 50/73 (68%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 151 LFLNNLPEETNEMMLSMLFNQFPGFKEVRLVPGRHDIAFVEFENETEAGSARDALQGFKI 210
Query 73 SSSHQLRISHVER 85
+ SH ++I++ ++
Sbjct 211 TPSHAMKITYAKK 223
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 41/82 (50%), Gaps = 8/82 (9%)
Query 8 LEARASLFVENLAEGTSKGALE----ILFSQYKGFREV---RLVEGRGVAFVDFAAPAAA 60
+ ++++ NL + K L+ LFSQ+ ++ + ++ RG AFV F ++A
Sbjct 3 IRPNHTVYINNLCDKVKKPELKRSLYALFSQFGHVVDIVALKTMKMRGQAFVIFKELSSA 62
Query 61 EVALSGLQGFKISSSHQLRISH 82
AL LQGF S +RI +
Sbjct 63 TNALRQLQGFPFYSK-PMRIQY 83
> ath:AT2G30260 U2B''; U2B'' (U2 small nuclear ribonucleoprotein
B); RNA binding / nucleic acid binding / nucleotide binding;
K11094 U2 small nuclear ribonucleoprotein B''
Length=232
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 28/73 (38%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF++NL T+ L++LF QY GF+E+R+++ + G+AFV++ A +A+ LQGFKI
Sbjct 160 LFIQNLPHETTSMMLQLLFEQYPGFKEIRMIDAKPGIAFVEYEDDVQASIAMQPLQGFKI 219
Query 73 SSSHQLRISHVER 85
+ + + IS ++
Sbjct 220 TPQNPMVISFAKK 232
> xla:495019 snrpb2; small nuclear ribonucleoprotein polypeptide
B; K11094 U2 small nuclear ribonucleoprotein B''
Length=223
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 151 LFLNNLPEETNEMMLSMLFNQFPGFKEVRLVPGRHDIAFVEFDNETEAGSARDALQGFKI 210
Query 73 SSSHQLRISHVER 85
+ S+ ++I+ ++
Sbjct 211 TPSNAMKITFAKK 223
> xla:379781 snrpa, MGC52976, U1A, snf; small nuclear ribonucleoprotein
polypeptide A; K11091 U1 small nuclear ribonucleoprotein
A
Length=282
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 34/73 (46%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 210 LFLTNLPEETNELMLSMLFNQFPGFKEVRLVPGRHDIAFVEFDNEVQAGAARESLQGFKI 269
Query 73 SSSHQLRISHVER 85
+ S+ ++IS ++
Sbjct 270 TQSNSMKISFAKK 282
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 7/65 (10%)
Query 13 SLFVENLAEGTSKGALE----ILFSQYKGFREV---RLVEGRGVAFVDFAAPAAAEVALS 65
++++ NL E K L+ +FSQ+ ++ R ++ RG AFV F ++A AL
Sbjct 11 TIYINNLNEKIKKDELKKSLYAIFSQFGQILDILVSRNLKMRGQAFVIFKETSSATNALR 70
Query 66 GLQGF 70
+QGF
Sbjct 71 SMQGF 75
> dre:324121 snrpa, fc19d01, wu:fc19d01, zgc:77810; small nuclear
ribonucleoprotein polypeptide A; K11091 U1 small nuclear
ribonucleoprotein A
Length=281
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 34/73 (46%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 209 LFLTNLPEETNELMLSMLFNQFPGFKEVRLVPGRHDIAFVEFDNEVQAGAAREALQGFKI 268
Query 73 SSSHQLRISHVER 85
+ S+ ++IS ++
Sbjct 269 TQSNAMKISFAKK 281
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 34/65 (52%), Gaps = 7/65 (10%)
Query 13 SLFVENLAEGTSKGALE----ILFSQYKGFREV---RLVEGRGVAFVDFAAPAAAEVALS 65
++++ NL E K L+ +FSQ+ ++ R ++ +G AFV F +A AL
Sbjct 11 TIYINNLNEKIKKDELKKSLYAIFSQFGQILDILVSRTLKMKGQAFVIFKEINSASNALR 70
Query 66 GLQGF 70
+QGF
Sbjct 71 SMQGF 75
> mmu:53607 Snrpa, C430021M15Rik, Rnu1a-1, Rnu1a1, U1A; small
nuclear ribonucleoprotein polypeptide A; K11091 U1 small nuclear
ribonucleoprotein A
Length=287
Score = 65.1 bits (157), Expect = 6e-11, Method: Composition-based stats.
Identities = 33/73 (45%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 215 LFLTNLPEETNELMLSMLFNQFPGFKEVRLVPGRHDIAFVEFDNEVQAGAARDALQGFKI 274
Query 73 SSSHQLRISHVER 85
+ ++ ++IS ++
Sbjct 275 TQNNAMKISFAKK 287
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 38/71 (53%), Gaps = 9/71 (12%)
Query 9 EARA--SLFVENLAEGTSKGALE----ILFSQYKGFREV---RLVEGRGVAFVDFAAPAA 59
E RA ++++ NL E K L+ +FSQ+ ++ R+++ RG AFV F +
Sbjct 11 ETRANHTIYINNLNEKIKKDELKKSLYAIFSQFGQILDILVSRIMKMRGQAFVIFKEVTS 70
Query 60 AEVALSGLQGF 70
A AL +QGF
Sbjct 71 ATNALRSMQGF 81
> ath:AT1G06960 small nuclear ribonucleoprotein U2B, putative
/ spliceosomal protein, putative
Length=200
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 50/82 (60%), Gaps = 1/82 (1%)
Query 5 KGALEARASLFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVA 63
+ + LF+ NL T+ L++LF QY GF+E+R++E + G+AFV++ + +A
Sbjct 119 QDTMPPNNILFIHNLPIETNSMMLQLLFEQYPGFKEIRMIEAKPGIAFVEYEDDVQSSMA 178
Query 64 LSGLQGFKISSSHQLRISHVER 85
+ LQGFKI+ + + +S ++
Sbjct 179 MQALQGFKITPQNPMVVSFAKK 200
> hsa:6626 SNRPA, Mud1, U1-A, U1A; small nuclear ribonucleoprotein
polypeptide A; K11091 U1 small nuclear ribonucleoprotein
A
Length=282
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 33/73 (45%), Positives = 49/73 (67%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR +AFV+F A A LQGFKI
Sbjct 210 LFLTNLPEETNELMLSMLFNQFPGFKEVRLVPGRHDIAFVEFDNEVQAGAARDALQGFKI 269
Query 73 SSSHQLRISHVER 85
+ ++ ++IS ++
Sbjct 270 TQNNAMKISFAKK 282
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 7/65 (10%)
Query 13 SLFVENLAEGTSKGALE----ILFSQYKGFREV---RLVEGRGVAFVDFAAPAAAEVALS 65
++++ NL E K L+ +FSQ+ ++ R ++ RG AFV F ++A AL
Sbjct 11 TIYINNLNEKIKKDELKKSLYAIFSQFGQILDILVSRSLKMRGQAFVIFKEVSSATNALR 70
Query 66 GLQGF 70
+QGF
Sbjct 71 SMQGF 75
> mmu:667350 Gm8587, EG667350; predicted pseudogene 8587
Length=309
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEG-RGVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+E LV G +AFV+F A AL LQGFKI
Sbjct 212 LFLTNLPEETNELMLSMLFTQFPGFKEAHLVPGCHDIAFVEFDNEVQAGAALDALQGFKI 271
Query 73 SSSHQLRISHVER 85
+ ++ ++IS V++
Sbjct 272 TQNNAMKISFVKK 284
> cel:K08D10.4 rnp-2; RNP (RRM RNA binding domain) containing
family member (rnp-2)
Length=206
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 47/81 (58%), Gaps = 1/81 (1%)
Query 6 GALEARASLFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVAL 64
G LF NL + + LEI+F+Q+ G +++R+V R G+AFV+F + A A
Sbjct 126 GPAPPNKILFCTNLPDSATAEMLEIMFNQFAGLKDIRMVPNRPGIAFVEFDTDSLAIPAR 185
Query 65 SGLQGFKISSSHQLRISHVER 85
+ L FKIS+ H +R+ + ++
Sbjct 186 TTLNNFKISAEHTMRVDYAKK 206
> mmu:382099 Gm5161, EG382099; predicted pseudogene 5161
Length=282
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 32/73 (43%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGR-GVAFVDFAAPAAAEVALSGLQGFKI 72
LF+ NL E T++ L +LF+Q+ GF+EVRLV GR + FV+F A A LQGFKI
Sbjct 210 LFLTNLPEETNELMLSMLFTQFPGFKEVRLVPGRHDIDFVEFDNEVQAGAARDVLQGFKI 269
Query 73 SSSHQLRISHVER 85
+ ++ ++IS ++
Sbjct 270 TQNNAMKISFAKK 282
> pfa:MAL13P1.35 U1 small nuclear ribonucleoprotein A, putative
Length=449
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 41/72 (56%), Gaps = 1/72 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKIS 73
LFVEN+ E A LF Y GF E R++ R VAFVDF A A+ +Q +++
Sbjct 379 LFVENVVENVDTQAFNDLFKNYAGFVEARIIPQRNVAFVDFTDETTATFAMKAVQNYELQ 438
Query 74 SSHQLRISHVER 85
S +L+IS+ +R
Sbjct 439 GS-KLKISYAKR 449
> cel:K08D10.3 rnp-3; RNP (RRM RNA binding domain) containing
family member (rnp-3)
Length=217
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEG-RGVAFVDFAAPAAAEVALSGLQGFKI 72
LF N+ EGT ++ +FSQ+ G REVR + + AF+++ + +E A L F+I
Sbjct 145 LFCSNIPEGTEPEQIQTIFSQFPGLREVRWMPNTKDFAFIEYESEDLSEPARQALDNFRI 204
Query 73 SSSHQLRI 80
+ + Q+ +
Sbjct 205 TPTQQITV 212
> ath:AT1G76940 RNA recognition motif (RRM)-containing protein
Length=233
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 40/76 (52%), Gaps = 9/76 (11%)
Query 9 EARASLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRG---------VAFVDFAAPAA 59
+A +L+VE L S+ + +F + G+REVRLV + FVDF PA
Sbjct 133 DASNTLYVEGLPSNCSRREVAHIFRPFVGYREVRLVTKDSKHRNGDPIVLCFVDFTNPAC 192
Query 60 AEVALSGLQGFKISSS 75
A ALS LQG+++ +
Sbjct 193 AATALSALQGYRMDEN 208
> bbo:BBOV_III004120 17.m07378; U2 small nuclear ribonucleoprotein
B'
Length=285
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 29/73 (39%), Positives = 42/73 (57%), Gaps = 1/73 (1%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKI 72
+LFVE L EG S + +F++ GF+E R++ R VAF+DF A A+ LQ +
Sbjct 214 TLFVEGLPEGVSLADVNAVFARSVGFQEARVIAARKVAFIDFDNEFNAGCAMQALQEHPM 273
Query 73 SSSHQLRISHVER 85
S LRIS+ +R
Sbjct 274 GDS-TLRISYAKR 285
> tgo:TGME49_109800 small nuclear ribonucleoprotein U1A, putative
Length=385
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 43/72 (59%), Gaps = 1/72 (1%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKIS 73
LF+ENL E + L LFS++ G EVR V R VAFV++ A A++ LQG+ ++
Sbjct 315 LFLENLPEDATMEGLVSLFSKHAGMIEVRPVLWRRVAFVEYDNEMLAANAMNALQGYNMN 374
Query 74 SSHQLRISHVER 85
S ++I++ R
Sbjct 375 GS-SIKITYARR 385
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 44/87 (50%), Gaps = 8/87 (9%)
Query 3 TSKGALEARASLFVENLAEGTSKGAL-EILFSQYKGFREVRLVEG------RGVAFVDFA 55
T ++ ++++ N+ + L E L S +K F E+R + RG A++ FA
Sbjct 105 TQDMSIPPNQTIYINNINDKVKLPELKENLRSMFKQFGEIRQIVAMSSFWRRGQAWIVFA 164
Query 56 APAAAEVALSGLQGFKISSSHQLRISH 82
+ +A A+ G+QGF + LRI++
Sbjct 165 SVESATKAIQGMQGF-VYHDQPLRINY 190
> cel:F46A9.6 mec-8; MEChanosensory abnormality family member
(mec-8)
Length=312
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 48/83 (57%), Gaps = 4/83 (4%)
Query 4 SKGALEARASLFVENLAEGTSKGALEILFSQYKGFREVRL--VEGRGVAFVDFAAPAAAE 61
S+ + A ++LFV NL+ ++ L +F + GF +RL G VAFV+++ A
Sbjct 220 SQASTSACSTLFVANLSAEVNEDTLRGVFKAFSGFTRLRLHNKNGSCVAFVEYSDLQKAT 279
Query 62 VALSGLQGFKISSSHQ--LRISH 82
A+ LQGF+I+++ + LRI +
Sbjct 280 QAMISLQGFQITANDRGGLRIEY 302
> ath:AT3G13700 RNA-binding protein, putative
Length=287
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 42/76 (55%), Gaps = 4/76 (5%)
Query 4 SKGALEARA--SLFVENLAEGTSKGALEILFSQYKGFR--EVRLVEGRGVAFVDFAAPAA 59
S+G ARA +LF+ NL ++ L+ L S+Y GF ++R G VAF DF
Sbjct 195 SEGGSGARACSTLFIANLGPNCTEDELKQLLSRYPGFHILKIRARGGMPVAFADFEEIEQ 254
Query 60 AEVALSGLQGFKISSS 75
A A++ LQG +SSS
Sbjct 255 ATDAMNHLQGNLLSSS 270
> ath:AT1G21320 nucleic acid binding / nucleotide binding
Length=253
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 36/70 (51%), Gaps = 9/70 (12%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLV---------EGRGVAFVDFAAPAAAEVA 63
+L+VE L S+ + +F + G+REVRLV + + FVDF A A A
Sbjct 156 TLYVEGLPSNCSRREVSHIFRPFVGYREVRLVTQDSKHRSGDPTVLCFVDFENSACAATA 215
Query 64 LSGLQGFKIS 73
LS LQ +++
Sbjct 216 LSALQDYRMD 225
> sce:YGR250C Putative RNA binding protein; localizes to stress
granules induced by glucose deprivation; interacts with Rbg1p
in a two-hybrid
Length=781
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 26/90 (28%), Positives = 42/90 (46%), Gaps = 6/90 (6%)
Query 2 GTSKGALEARASLFVENLAEGTSKGALEILFSQYKGFREVRLV------EGRGVAFVDFA 55
G +K AL ++FV +A+ S G L LFS+Y ++L+ E G F+ +
Sbjct 185 GVNKHALTYPGNIFVGGIAKSLSIGELSFLFSKYGPILSMKLIYDKTKGEPNGYGFISYP 244
Query 56 APAAAEVALSGLQGFKISSSHQLRISHVER 85
+ A + + L G ++ S HVER
Sbjct 245 LGSQASLCIKELNGRTVNGSTLFINYHVER 274
> sce:YBR212W NGR1, RBP1; Ngr1p
Length=672
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 35/69 (50%), Gaps = 1/69 (1%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKI 72
++FV L T++ L LF + VR+ G+ FV F AE ++ GLQGF +
Sbjct 361 TVFVGGLVPKTTEFQLRSLFKPFGPILNVRIPNGKNCGFVKFEKRIDAEASIQGLQGFIV 420
Query 73 SSSHQLRIS 81
S +R+S
Sbjct 421 GGS-PIRLS 428
> mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=380
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + ++ LE LFSQY R+ V G RGV F+ F AE A+
Sbjct 132 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIK 191
Query 66 GLQGFKISSSHQ 77
GL G K S + +
Sbjct 192 GLNGQKPSGATE 203
> tpv:TP03_0036 hypothetical protein
Length=288
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDF 54
LFVE+L +G + + LF GF E +L+ + VAF+DF
Sbjct 205 LFVEDLPDGIAHEDVNNLFHHMPGFVETKLINSKHVAFIDF 245
> hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV like
protein 2/3/4
Length=366
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + ++ LE LFSQY R+ V G RGV F+ F AE A+
Sbjct 132 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIK 191
Query 66 GLQGFKISSSHQ 77
GL G K S + +
Sbjct 192 GLNGQKPSGATE 203
> xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic
lethal, abnormal vision)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=400
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + ++ LE LFSQY R+ V G RGV F+ F AE A+
Sbjct 166 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIK 225
Query 66 GLQGFKISSSHQ 77
GL G K S + +
Sbjct 226 GLNGQKPSGAAE 237
> ath:AT3G16380 PAB6; PAB6 (POLY(A) BINDING PROTEIN 6); RNA binding
/ translation initiation factor
Length=537
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 5/67 (7%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLVE-----GRGVAFVDFAAPAAAEVALSGL 67
+++V+NL E + L LFSQY V ++ RG FV+F P A+ A+ L
Sbjct 203 NVYVKNLIETVTDDCLHTLFSQYGTVSSVVVMRDGMGRSRGFGFVNFCNPENAKKAMESL 262
Query 68 QGFKISS 74
G ++ S
Sbjct 263 CGLQLGS 269
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 4/62 (6%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRLVE----GRGVAFVDFAAPAAAEVALSGL 67
A+L+V+NL + LE +F + ++VE +G FV F +A A S L
Sbjct 112 ANLYVKNLDSSITSSCLERMFCPFGSILSCKVVEENGQSKGFGFVQFDTEQSAVSARSAL 171
Query 68 QG 69
G
Sbjct 172 HG 173
> mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 2 (Hu antigen B); K13208 ELAV like
protein 2/3/4
Length=359
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + ++ LE LFSQY R+ V G RGV F+ F AE A+
Sbjct 125 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGISRGVGFIRFDKRIEAEEAIK 184
Query 66 GLQGFK 71
GL G K
Sbjct 185 GLNGQK 190
> hsa:1993 ELAVL2, HEL-N1, HELN1, HUB; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 2 (Hu antigen B); K13208
ELAV like protein 2/3/4
Length=346
Score = 37.4 bits (85), Expect = 0.013, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + ++ LE LFSQY R+ V G RGV F+ F AE A+
Sbjct 125 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGISRGVGFIRFDKRIEAEEAIK 184
Query 66 GLQGFK 71
GL G K
Sbjct 185 GLNGQK 190
> dre:321994 nono, wu:fb40d10, wu:fc11d05; non-POU domain containing,
octamer-binding; K13214 non-POU domain-containing octamer-binding
protein
Length=441
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Query 11 RASLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGF 70
R+ LFV NL GT++ +E LFS+Y E+ + + RG F+ A++A + L
Sbjct 71 RSRLFVGNLPAGTTEEDVEKLFSKYGKASEIFINKDRGFGFIRLETKTLADIAKAELDD- 129
Query 71 KISSSHQLRI 80
I Q+R+
Sbjct 130 TIFRGRQIRV 139
> xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,
hub, p45; ELAV (embryonic lethal, abnormal vision)-like 2
(Hu antigen B); K13208 ELAV like protein 2/3/4
Length=389
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + ++ LE LFSQY R+ V G RGV F+ F AE A+
Sbjct 153 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIK 212
Query 66 GLQGFK 71
GL G K
Sbjct 213 GLNGQK 218
> ath:AT2G42240 nucleic acid binding / nucleotide binding
Length=302
Score = 36.6 bits (83), Expect = 0.017, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 37/65 (56%), Gaps = 2/65 (3%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRL--VEGRGVAFVDFAAPAAAEVALSGLQGF 70
+LF+ N+ ++ L +FS+ +GF ++++ G VAFVDF + + AL LQG
Sbjct 206 TLFIANMGPNCTEAELIQVFSRCRGFLKLKIQGTYGTPVAFVDFQDVSCSSEALHTLQGT 265
Query 71 KISSS 75
+ SS
Sbjct 266 VLYSS 270
> xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1;
ELAV (embryonic lethal, abnormal vision)-like 2 (Hu antigen
B); K13208 ELAV like protein 2/3/4
Length=389
Score = 36.6 bits (83), Expect = 0.017, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + ++ LE LFSQY R+ V G RGV F+ F AE A+
Sbjct 153 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIK 212
Query 66 GLQGFK 71
GL G K
Sbjct 213 GLNGQK 218
> hsa:5935 RBM3, IS1-RNPL, RNPL; RNA binding motif (RNP1, RRM)
protein 3; K13186 RNA-binding protein 3
Length=157
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 7/75 (9%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVE------GRGVAFVDFAAPAAAEVALSGL 67
LFV L T + ALE FS + EV +V+ RG F+ F P A VA+ +
Sbjct 8 LFVGGLNFNTDEQALEDHFSSFGPISEVVVVKDRETQRSRGFGFITFTNPEHASVAMRAM 67
Query 68 QGFKISSSHQLRISH 82
G + Q+R+ H
Sbjct 68 NGESL-DGRQIRVDH 81
> dre:30737 elavl4, elrd, wu:fc24h01, zHuD; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 4 (Hu antigen D); K13208
ELAV like protein 2/3/4
Length=367
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 36/75 (48%), Gaps = 9/75 (12%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRLV---------EGRGVAFVDFAAPAAAEV 62
A+L+V L + ++ LE LFSQY R++ RGV F+ F AE
Sbjct 130 ANLYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGPTGGSRGVGFIRFDKRIEAEE 189
Query 63 ALSGLQGFKISSSHQ 77
A+ GL G K S + +
Sbjct 190 AIKGLNGQKPSGAAE 204
> cel:T23F6.4 rbd-1; RBD (RNA binding domain) protein family member
(rbd-1); K14787 multiple RNA-binding domain-containing
protein 1
Length=872
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 46/91 (50%), Gaps = 14/91 (15%)
Query 8 LEARASLFVENLAEGTSKGALEILFSQYKG--------FREVRLVE-----GRGVAFVDF 54
+E+ ++LFV+NLA T+ G+LE LF + G +++ E G FV F
Sbjct 637 IESGSTLFVKNLAFDTTDGSLEFLFRKRYGDLVKSAQISKKLNPAEPTKPLSMGFGFVQF 696
Query 55 AAPAAAEVALSGLQGFKIS-SSHQLRISHVE 84
A+ AL +QG + S +L+ISH E
Sbjct 697 YTAFDAKTALKDMQGELLDGHSLELKISHRE 727
> sce:YHR086W NAM8, MRE2, MUD15; Nam8p
Length=523
Score = 36.2 bits (82), Expect = 0.025, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 39/69 (56%), Gaps = 1/69 (1%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKI 72
++F+ L+ ++ L F + V++ G+ FV + +AE A++G+QGF I
Sbjct 314 TVFIGGLSSLVTEDELRAYFQPFGTIVYVKIPVGKCCGFVQYVDRLSAEAAIAGMQGFPI 373
Query 73 SSSHQLRIS 81
++S ++R+S
Sbjct 374 ANS-RVRLS 381
> ath:AT5G47320 RPS19; RPS19 (RIBOSOMAL PROTEIN S19); RNA binding;
K02965 small subunit ribosomal protein S19
Length=212
Score = 35.8 bits (81), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 11/71 (15%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLVEG------RGVAFVDFAAPAAAEVALSG- 66
L++ L+ GT + +L+ FS + G E R++ RG FV+F + +A A+S
Sbjct 33 LYIGGLSPGTDEHSLKDAFSSFNGVTEARVMTNKVTGRSRGYGFVNFISEDSANSAISAM 92
Query 67 ----LQGFKIS 73
L GF IS
Sbjct 93 NGQELNGFNIS 103
> cel:C07A4.1 hypothetical protein
Length=360
Score = 35.8 bits (81), Expect = 0.031, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 13 SLFVENLAEGTSKGALEILFSQYKGFREVRLVEGRGVAFVDFAAPAAAEVALSGLQGFKI 72
S++V N+++ T+ L LFS Y EVR+ + + AFV + A A+ + G K
Sbjct 238 SVYVGNISQQTTDADLRDLFSTYGDIAEVRIFKTQRYAFVRYEKKECATKAIMEMNG-KE 296
Query 73 SSSHQLRIS 81
+ +Q+R S
Sbjct 297 MAGNQVRCS 305
> hsa:1995 ELAVL3, DKFZp547J036, HUC, HUCL, MGC20653, PLE21; ELAV
(embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu
antigen C); K13208 ELAV like protein 2/3/4
Length=367
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 33/66 (50%), Gaps = 6/66 (9%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRLV------EGRGVAFVDFAAPAAAEVALS 65
A+L+V L + S+ +E LFSQY R++ RGV F+ F AE A+
Sbjct 125 ANLYVSGLPKTMSQKEMEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIK 184
Query 66 GLQGFK 71
GL G K
Sbjct 185 GLNGQK 190
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 18/79 (22%), Positives = 37/79 (46%), Gaps = 6/79 (7%)
Query 2 GTSKGALEARASLFVENLAEGTSKGALEILFSQYKGFREVRLVEGR------GVAFVDFA 55
GT+ +++ +L V L + ++ + LF +LV + G FV+++
Sbjct 29 GTNGATDDSKTNLIVNYLPQNMTQDEFKSLFGSIGDIESCKLVRDKITGQSLGYGFVNYS 88
Query 56 APAAAEVALSGLQGFKISS 74
P A+ A++ L G K+ +
Sbjct 89 DPNDADKAINTLNGLKLQT 107
> hsa:494115 RBMXL1, DKFZp547N1117, DKFZp667D0223, FLJ32614, RBM1;
RNA binding motif protein, X-linked-like 1; K12885 heterogeneous
nuclear ribonucleoprotein G
Length=390
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 33/62 (53%), Gaps = 6/62 (9%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLV------EGRGVAFVDFAAPAAAEVALSGL 67
LF+ L T++ ALE +F +Y EV L+ + RG AFV F +PA A+ A +
Sbjct 10 LFIGGLNTETNEKALETVFGKYGRIVEVLLIKDRETNKSRGFAFVTFESPADAKDAARDM 69
Query 68 QG 69
G
Sbjct 70 NG 71
> ath:AT5G07290 AML4; AML4 (ARABIDOPSIS MEI2-LIKE 4); RNA binding
/ nucleic acid binding / nucleotide binding
Length=907
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 38/73 (52%), Gaps = 2/73 (2%)
Query 10 ARASLFVENLAEGTSKGALEILFSQYKGFREV-RLVEGRGVAFVDFAAPAAAEVALSGLQ 68
+ +L+V NL S L +FS Y REV R + +++F A+VAL GL
Sbjct 293 SEGALWVNNLDSSISNEELHGIFSSYGEIREVRRTMHENSQVYIEFFDVRKAKVALQGLN 352
Query 69 GFKISSSHQLRIS 81
G ++ + QL+++
Sbjct 353 GLEV-AGRQLKLA 364
> pfa:PFI1025w RNA binding protein, putative
Length=480
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 36/76 (47%), Gaps = 5/76 (6%)
Query 14 LFVENLAEGTSKGALEILFSQYKGFREVRLV-----EGRGVAFVDFAAPAAAEVALSGLQ 68
L+V+N+ E +K E FS+ G+ E R V R A++DF A L LQ
Sbjct 114 LYVKNINEHITKENFESYFSKIDGYIETRFVVDTSRTSRKFAYIDFENKTKALAFLHTLQ 173
Query 69 GFKISSSHQLRISHVE 84
I + L +++V+
Sbjct 174 NSNIENFKTLTLNNVD 189
> dre:30732 elavl3, HuC, elrc, id:ibd1248, wu:fb77b03, zHuC; ELAV
(embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu
antigen C); K13208 ELAV like protein 2/3/4
Length=345
Score = 35.8 bits (81), Expect = 0.033, Method: Composition-based stats.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 6/66 (9%)
Query 12 ASLFVENLAEGTSKGALEILFSQYKGFREVRL----VEG--RGVAFVDFAAPAAAEVALS 65
A+L+V L + S+ +E LFSQY R+ V G RGV F+ F AE A+
Sbjct 124 ANLYVSGLPKTMSQKDMEQLFSQYGRIITSRILVNQVTGISRGVGFIRFDKRNEAEEAIK 183
Query 66 GLQGFK 71
GL G K
Sbjct 184 GLNGQK 189
Lambda K H
0.317 0.133 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2035463248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40